RNAseq tutorial without coding
Marie Saitou
5/26/2022
Last updated: 2022-05-29
Checks: 6 1
Knit directory: Bio326/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210128) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d86965b. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .RData
Ignored: .Rhistory
Ignored: analysis/.DS_Store
Ignored: analysis/popgen.simu.nb.html
Untracked files:
Untracked: BIO326 URL genome annotatin computer lab_24_MAR_2021.docx
Untracked: BIO326-121VGenomesequencingBIO326-121VGenomsekvensering;verktøyoganalyser-BIO326-121VGenomesequencing_PhillipByronPope.pdf
Untracked: BIO326-RNAseq.pptx
Untracked: BIO326-genome/
Untracked: BIO326.MS.10th_FEB_2021function.pptx
Untracked: BIO326_Introduction to sequence technology and protocols_3rd_FEB_2021.pdf
Untracked: BIO326_Introduction to sequence technology and protocols_3rd_FEB_2021.pptx
Untracked: BIO326_RNAseq_5th_FEB_2021.pptx
Untracked: BIO326_SQK-RAD004 DNA challenge.docx
Untracked: BIO326_visual_30_APR_2021.pptx
Untracked: Bio326.2022.1.Rmd
Untracked: Bio326.genome.html
Untracked: Nanopore_SumStatQC_Tutorial.Rmd
Untracked: PCRdemo.R
Untracked: Pig_mutation_hist.csv
Untracked: PopGenBio326.322/
Untracked: RNAseq.Rplot.pdf
Untracked: RNAseq_Jun_2022.pdf
Untracked: RNAseq_Jun_2022.pptx
Untracked: Untitled.R
Untracked: [eng]BIO326-121VGenomesequencingBIO326-121VGenomsekvensering;verktøyoganalyser-BIO326-121VGenomesequencing_PhillipByronPope.mht
Untracked: [eng]BIO326-121VGenomesequencingBIO326-121VGenomsekvensering;verktøyoganalyser-BIO326-121VGenomesequencing_PhillipByronPope.pdf
Untracked: analysis/AnimalGenomics.Rmd
Untracked: analysis/AnimalGenomicsVariant2022.Rmd
Untracked: prepare.txt
Untracked: samples.xlsx
Untracked: test/
Untracked: trial/
Untracked: vis.xlsx
Untracked: workflowR.bio326.R
Unstaged changes:
Modified: analysis/RNAseq_for_lab.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/RNAseq_for_lab.Rmd) and HTML (docs/RNAseq_for_lab.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | d86965b | mariesaitou | 2022-05-26 | Build site. |
| Rmd | 617766e | mariesaitou | 2022-05-26 | wflow_publish(c(“analysis/RNAseq_for_lab.Rmd”)) |
| html | 88be94a | mariesaitou | 2022-05-26 | Build site. |
| Rmd | 2908374 | mariesaitou | 2022-05-26 | wflow_publish(c(“analysis/RNAseq_for_lab.Rmd”)) |
| html | ee6ef1d | mariesaitou | 2022-05-26 | Build site. |
| Rmd | 0c312c3 | mariesaitou | 2022-05-26 | image data set |
| html | 0c312c3 | mariesaitou | 2022-05-26 | image data set |
| html | 4d3c3e5 | mariesaitou | 2022-05-26 | Build site. |
| Rmd | e5d285b | mariesaitou | 2022-05-26 | wflow_publish(c(“analysis/RNAseq_for_lab.Rmd”)) |
| html | 2084e7f | mariesaitou | 2022-05-26 | Build site. |
| Rmd | e4f0b6b | mariesaitou | 2022-05-26 | wflow_publish(c(“analysis/RNAseq_for_lab.Rmd”)) |
Lecture slides
Marie: Overview of the RNAseq. What can RNAseq tell us/What should we do when we write a manuscript. slides
Matthew: RNAseq lab part mechanism, experimental design etc. (coming soon)
Domniki: How to upload data to ENA. (coming soon)
0. Goal of this workflow
Objective: Conduct comparative RNA sequencing analyis between fish brain and liver datasets
We will learn:
A. How to conduct “cleaning” of the RNAseq data.
B. How to quantify the gene expression in each sample.
C. How to conduct downstream analysis to gain biological insights.
1. Galaxy introduction
Useful materials:
https://training.galaxyproject.org/training-material/topics/introduction/
1-1. Register and login to Galaxy
Go to https://usegalaxy.no/ ,
Galaxy is a web platform with various software for genome analyses.
You should be able to log in with “Feide” information at galaxy.no (NMBU ID and password)
If you are not NMBU employee, you can use these for free: https://usegalaxy.eu/ or https://usegalaxy.org/
1-2. Play around with Galaxy
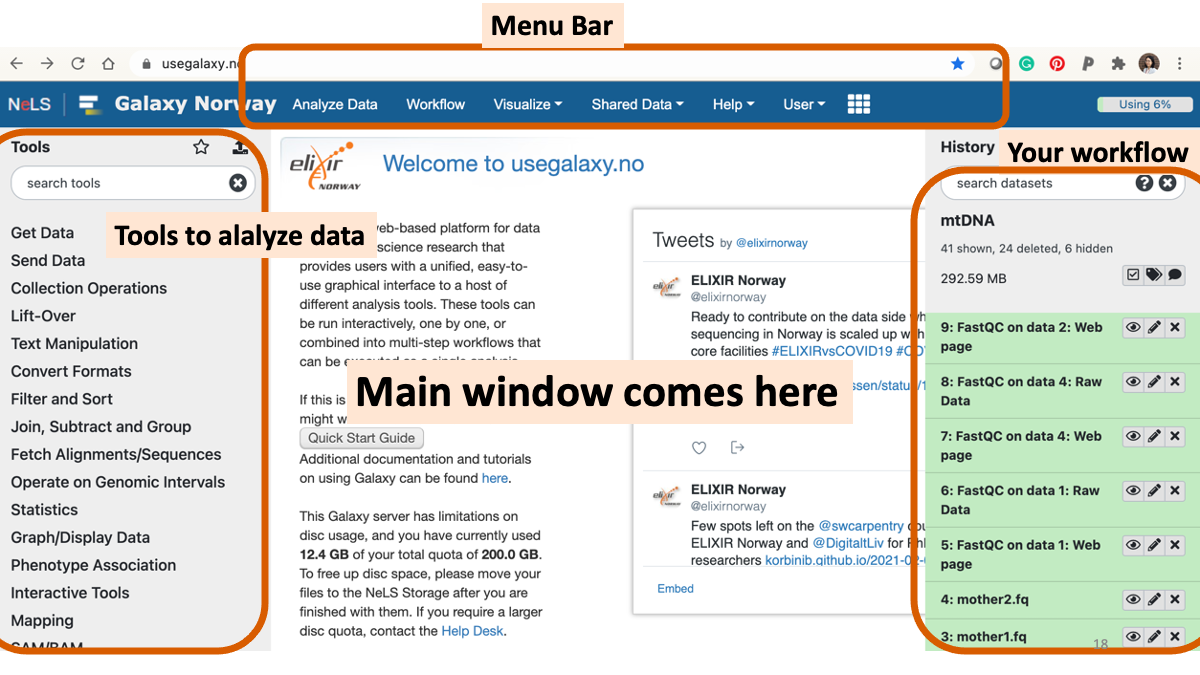
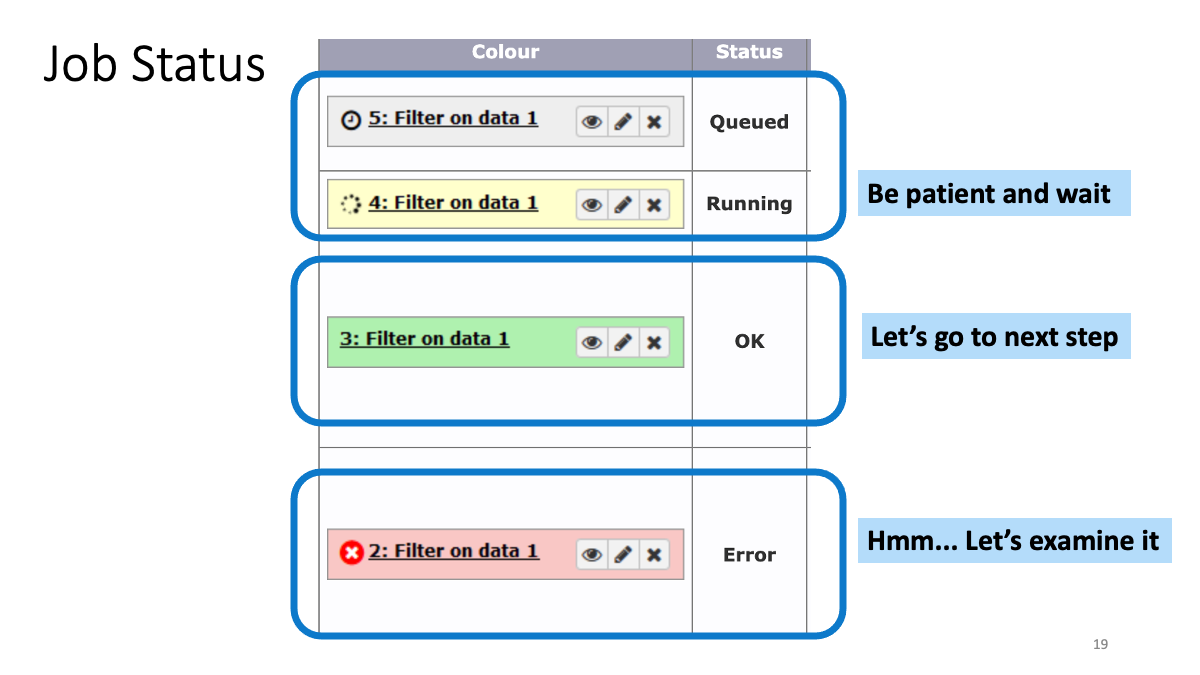


1-3. Review the RNAseq analysis workflow
On Galaxy
- Get RNAseq data (paired end, liver and brain, three samples each)
- Get reference transcriptome: Salmo salar version 3.1
- fastp: trimming low-quality reads
- Kallisto: quantify gene expression from the RNAseq data and reference
- tximport: summarize transcripts into genes
On iDEP
- Upload gene expression table
- Quality check
- Plot genes of interest
- Differentially expressed gene
- Gene ontology analysis2. Get the data
There are three major public repository for genomics data ENA (Europe), NCBI (America) DDBJ (Japan) – which are regularly synchronized.
This time, we will analyze brain and liver transcriptome datasets from the following study: “Multi-tissue transcriptome profiling of North American derived Atlantic salmon”
Paper: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6146974/
Dataset: https://www.ebi.ac.uk/ena/browser/view/PRJNA470665
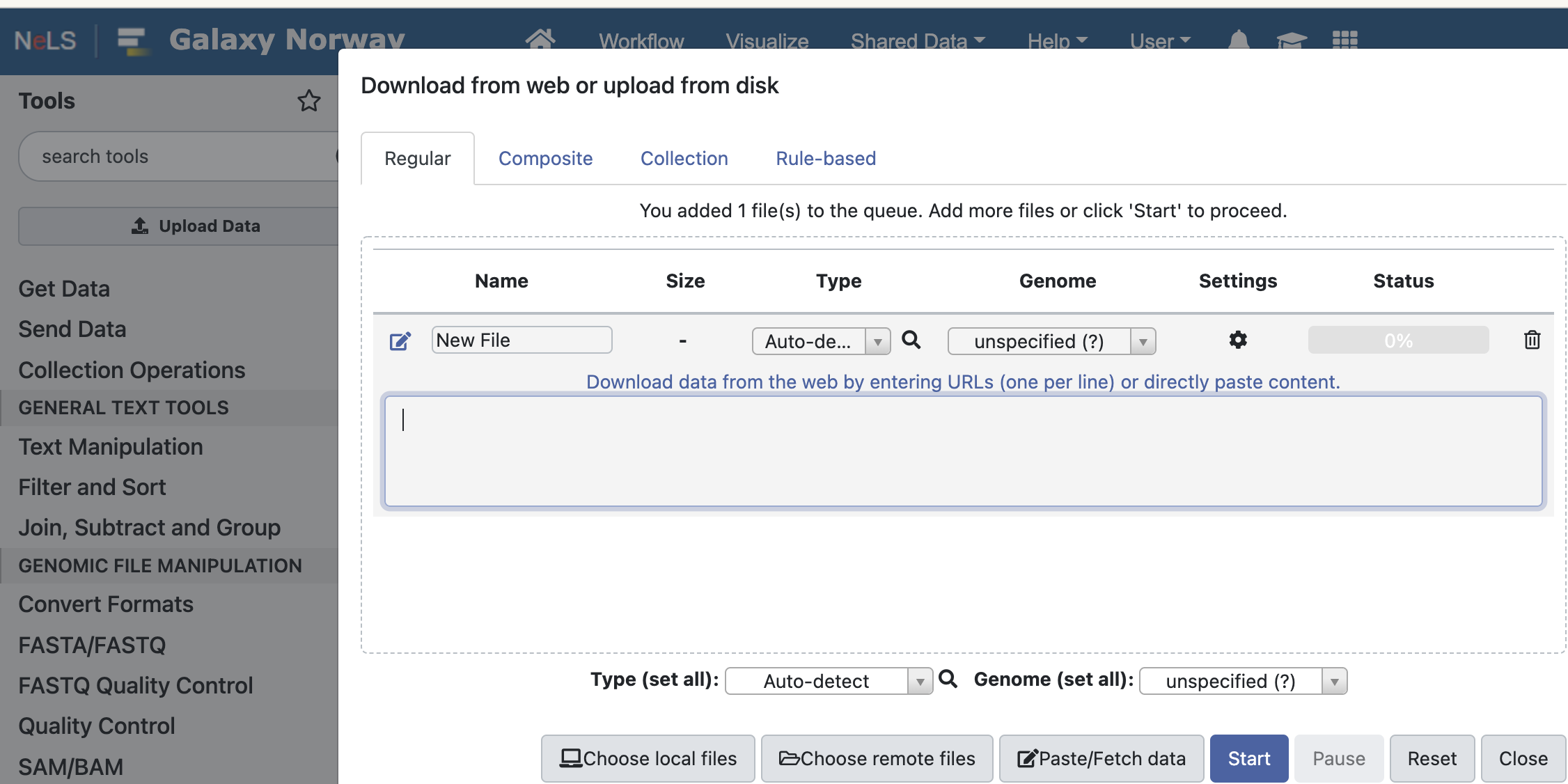 Click “upload data” on the left top - and
Click “upload data” on the left top - and
Select Paste/Fetch data and copy/paste the URL of the datasets.
Choose local files — upload data from your computer.
Choose remote files — if your Galaxy storage (200Gb in total) is getting full, you can store large files at the NeLS Storage and access them here.
To transfer files to the NeLS Storage, click Send Data - Export datasets in the left menu and select NeLS Storage as the destination. 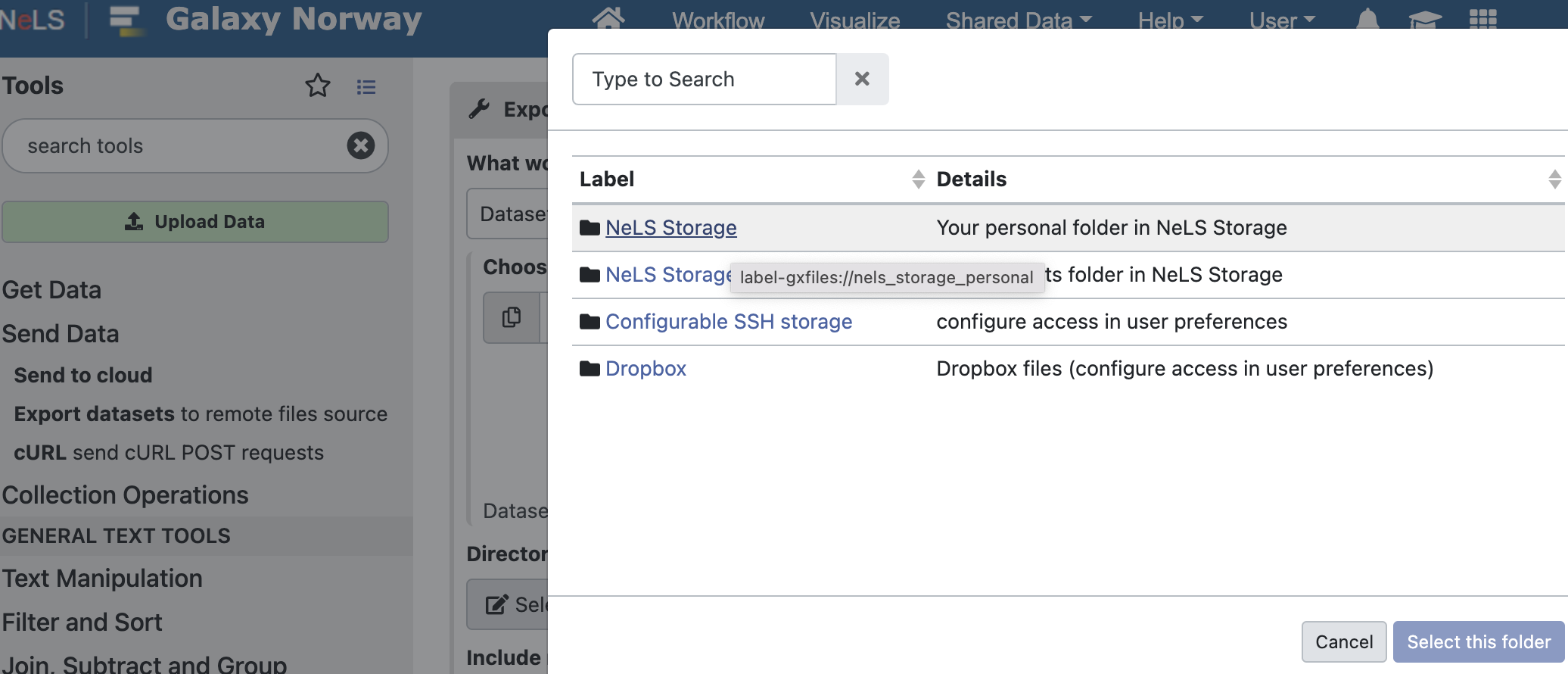
2-1. Get the liver and brain RNAseq data
As it takes hours to analyze the real (gigabytes) data, I made a miniature datasets for each samples.
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139945_1_mini.fastq.gz
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139945_2_mini.fastq.gz
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139950_1_mini.fastq.gz
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139950_2_mini.fastq.gz
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139969_1_mini.fastq.gz
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139969_2_mini.fastq.gz
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139971_1_mini.fastq.gz
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139971_2_mini.fastq.gz
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139973_1_mini.fastq.gz
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139973_2_mini.fastq.gz
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139947_1_mini.fastq.gz
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139947_2_mini.fastq.gz
2-2. Get the reference
http://ftp.ensembl.org/pub/release-106/fasta/salmo_salar/cdna/Salmo_salar.Ssal_v3.1.cdna.all.fa.gz
## New! Hien newly made a chromosomes + mitochondrial genome version
http://arken.nmbu.no/~lagr/share/Ssal.withMT/ensembl/Salmo_salar.Ssal_v3.1.106.withMT.gff3.gz(If it does not work well, download it on your computer and upload it to Galaxy afterword)
If you need reference sequences of other species for your study, go to https://www.ensembl.org/index.html
and “select a species” in the all genomes tub.
Go to “Gene annotation”, Download FASTA files and cDNA sequence.
If you want to try with real data later, click here
# Real brain samples (two files per sample)
ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR713/005/SRR7139945/SRR7139945_1.fastq.gz
ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR713/005/SRR7139945/SRR7139945_2.fastq.gz
ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR713/007/SRR7139947/SRR7139947_1.fastq.gz
ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR713/007/SRR7139947/SRR7139947_2.fastq.gz
ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR713/000/SRR7139950/SRR7139950_1.fastq.gz
ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR713/000/SRR7139950/SRR7139950_2.fastq.gz
# Real liver samoles (two files per sample)
ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR713/009/SRR7139969/SRR7139969_1.fastq.gz
ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR713/009/SRR7139969/SRR7139969_2.fastq.gz
ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR713/001/SRR7139971/SRR7139971_1.fastq.gz
ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR713/001/SRR7139971/SRR7139971_2.fastq.gz
ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR713/003/SRR7139973/SRR7139973_1.fastq.gz
ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR713/003/SRR7139973/SRR7139973_2.fastq.gz
Marie’s note to make tutorial files
for file in *.fastq.gz; do zcat < $file | head -100000 | gzip > {$file}_mini.fastq.gz; done3. Quality Control
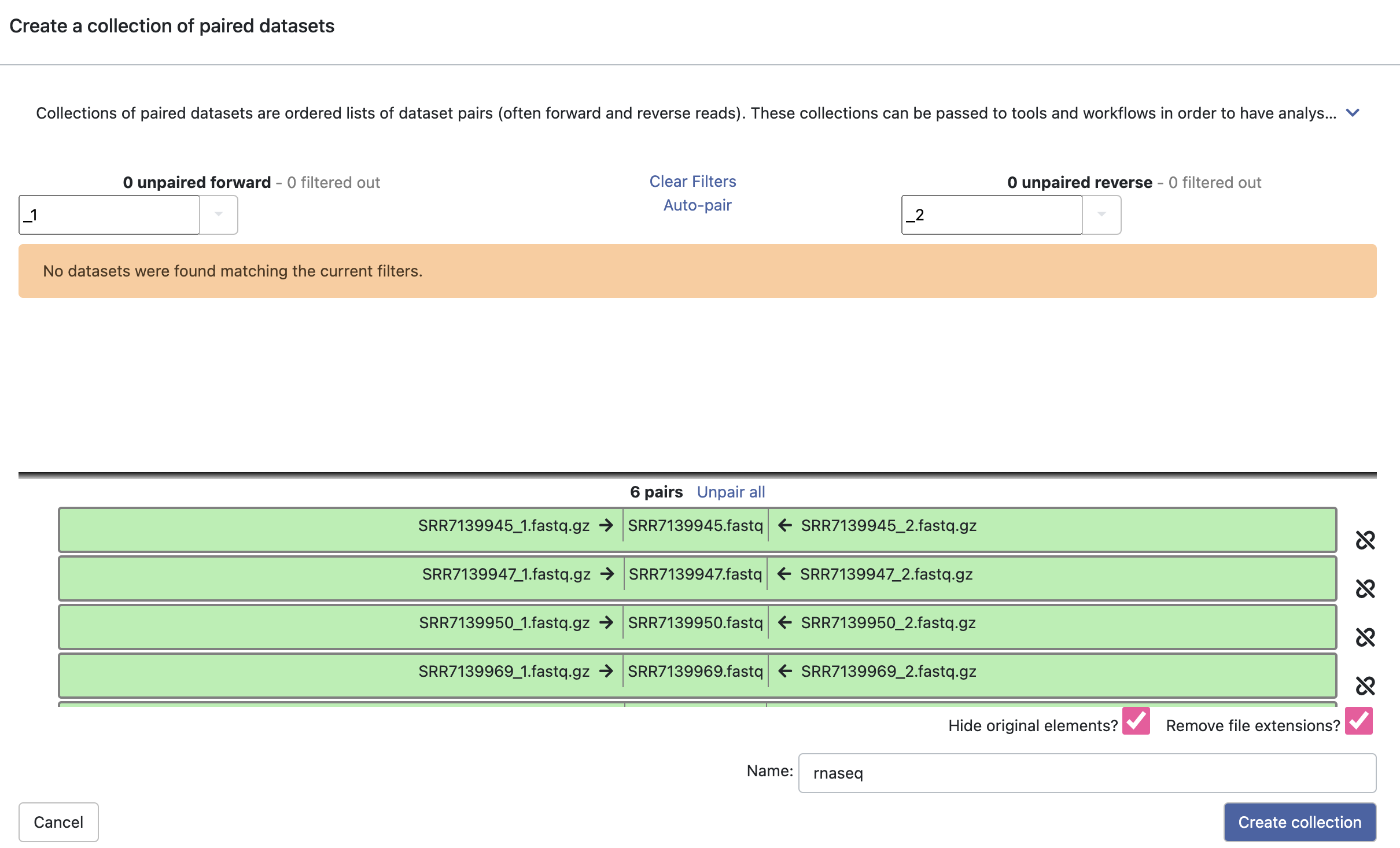
3-1. Pairing of the paired read data sets
What is Paired-End Sequencing?
As each sample has two files (paired-end), we make a “collection of paired datasets” to handle many files conveniently.
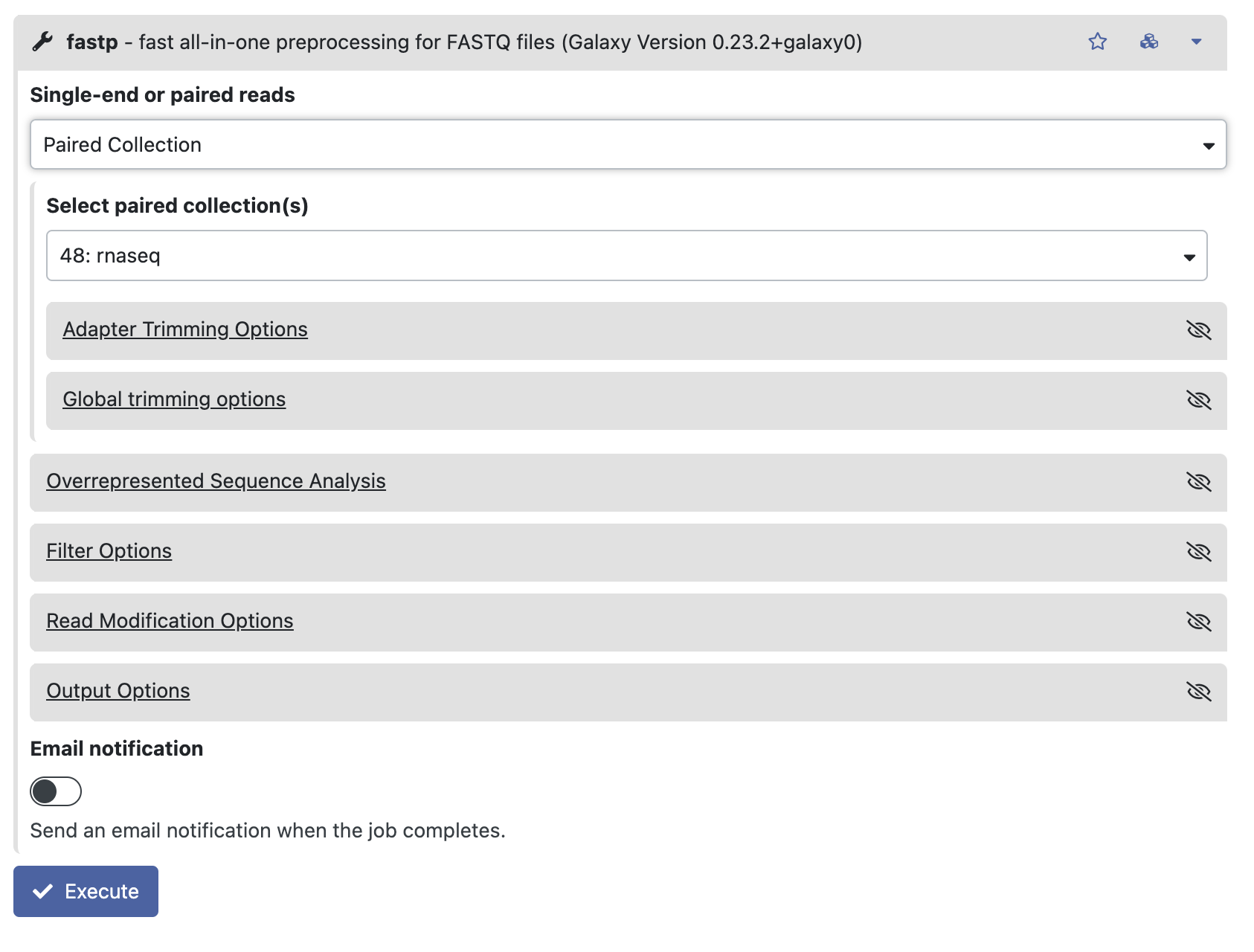
3-2. Trimming of bad quality data and adaptors with fastp
What fastp does (from Galaxy page)
fastp is a tool designed to provide fast all-in-one preprocessing for FASTQ files.
Features
- Filter out bad reads (too low quality, too short, or too many N…)
- Cut low quality bases for per read in its 5’ and 3’ by evaluating the mean quality from a sliding window (like Trimmomatic but faster) Trim all reads in front and tail
- Cut adapters. Adapter sequences can be automatically detected, which means you don’t have to input the adapter sequences to trim them.
- Correct mismatched base pairs in overlapped regions of paired end reads, if one base is with high quality while the other is with ultra-low quality
- Trim polyG in 3’ ends, which is commonly seen in NovaSeq/NextSeq data. Trim polyX in 3’ ends to remove unwanted polyX tailing (i.e. polyA tailing for mRNA-Seq data)
- Preprocess unique molecular identifer (UMI) enabled data, shift UMI to sequence name
- Report JSON format result for further interpreting
- Visualize quality control and filtering results on a single HTML page (like FASTQC but faster and more informative)
- Split the output to multiple files (0001.R1.gz, 0002.R1.gz…) to support parallel processing. Two modes can be used, limiting the total split file number, or limitting the lines of each split file (Not enabled in this Galaxy tool)
- Support long reads (data from PacBio / Nanopore devices)
Search for a tool “fastp” in the left window (Tools) and put the input files we gathered in the 3-1.
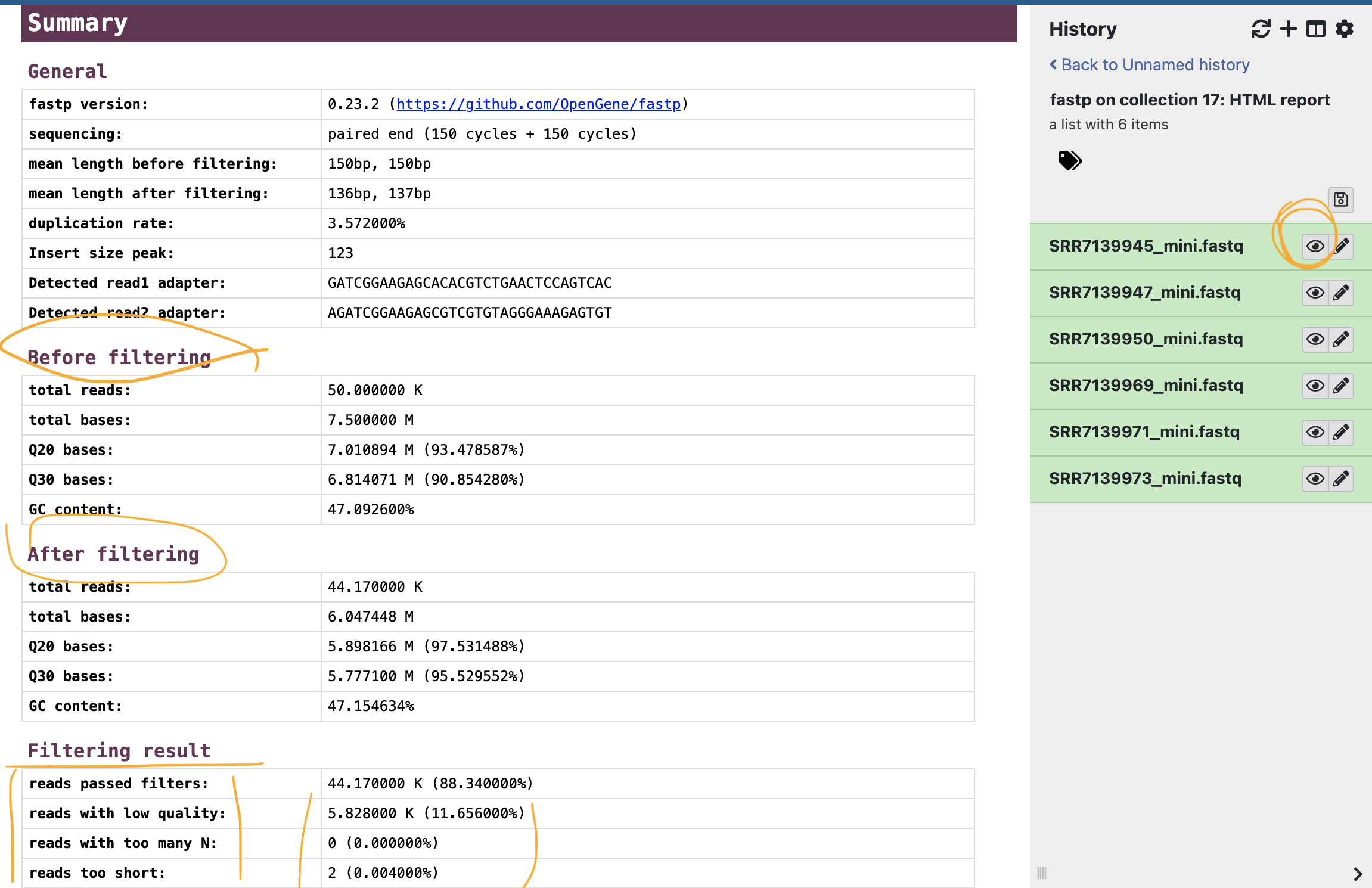
Let’s take a look on how much/what kind of reads were removed.
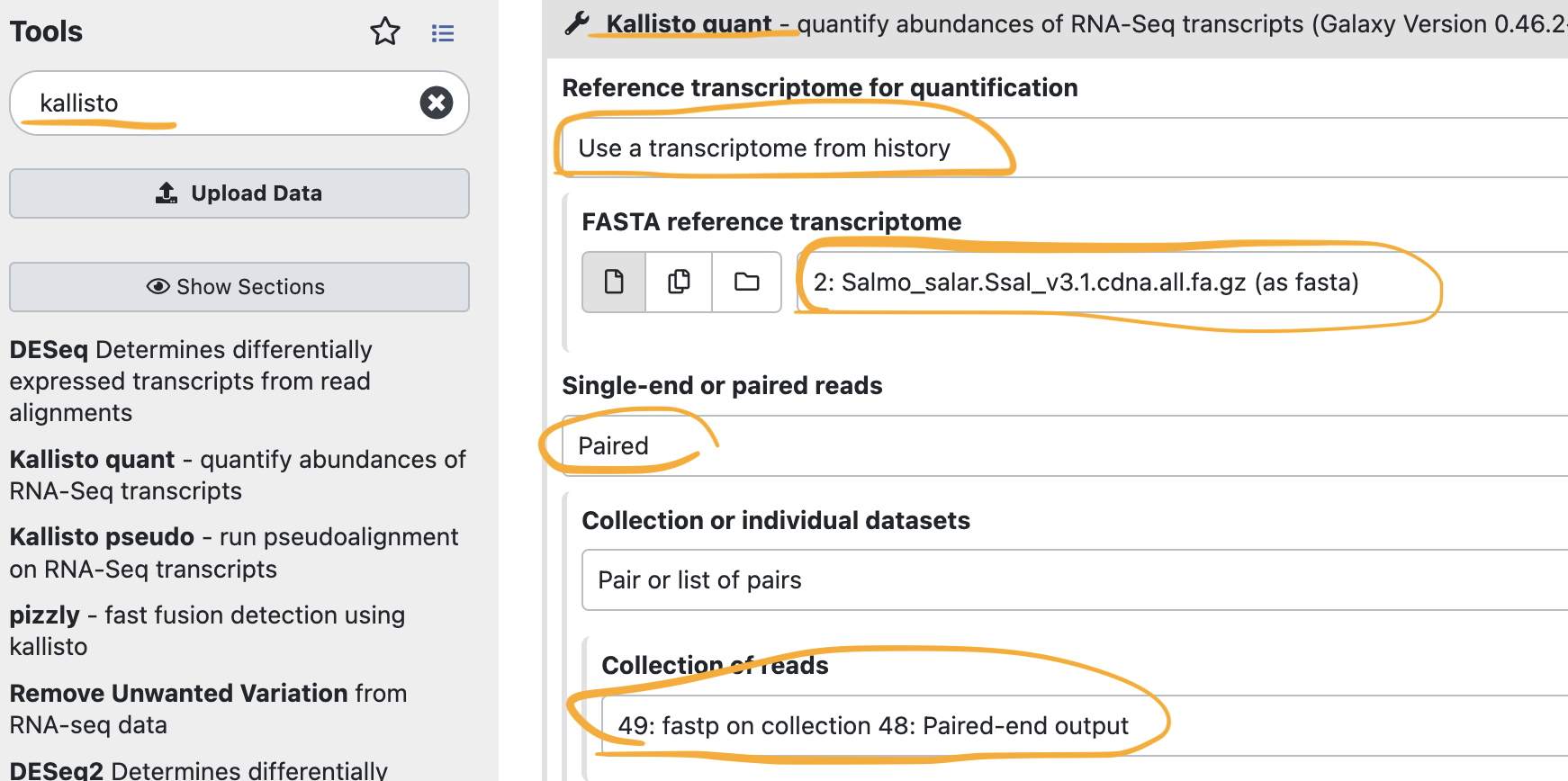
3-3. Quantify the gene expression on the cleaned reads with Kallisto
What Kallisto does (from Galaxy page)
kallisto is a program for quantifying abundances of transcripts from RNA-Seq data, or more generally of target sequences using high-throughput sequencing reads. It is based on the novel idea of pseudoalignment for rapidly determining the compatibility of reads with targets, without the need for alignment. On benchmarks with standard RNA-Seq data, kallisto can quantify 30 million human reads in less than 3 minutes on a Mac desktop computer using only the read sequences and a transcriptome index that itself takes less than 10 minutes to build. Pseudoalignment of reads preserves the key information needed for quantification, and kallisto is therefore not only fast, but also as accurate as existing quantification tools. In fact, because the pseudoalignment procedure is robust to errors in the reads, in many benchmarks kallisto significantly outperforms existing tools.
If it is taking too long, here are the output files. The output files are six in total, transcript * transcript counts tables.
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139945_mini.Kallisto.tabular
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139947_mini.Kallisto.tabular
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139969_mini.Kallisto.tabular
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139971_mini.Kallisto.tabular
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139973_mini.Kallisto.tabular
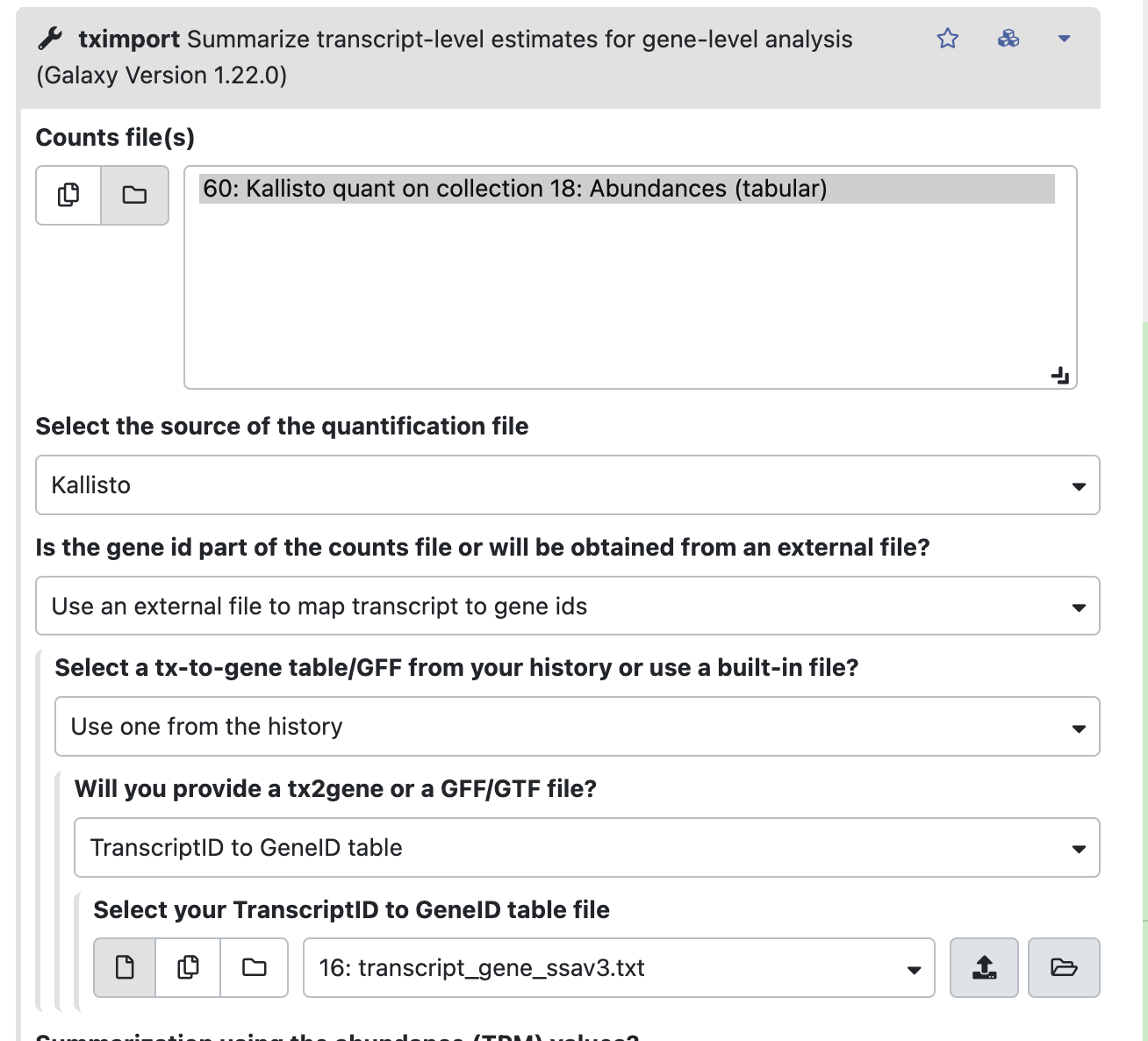
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/SRR7139950_mini.Kallisto.tabular3-4. Summarize the transcripts to genes with tximport
Upload a transcript-gene table to Galaxy.
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/transcript_gene_ssav3.txt(This table can be obtained at Ensemnbl BiomaRt for many species.)
4. Downstream analysis with iDEP
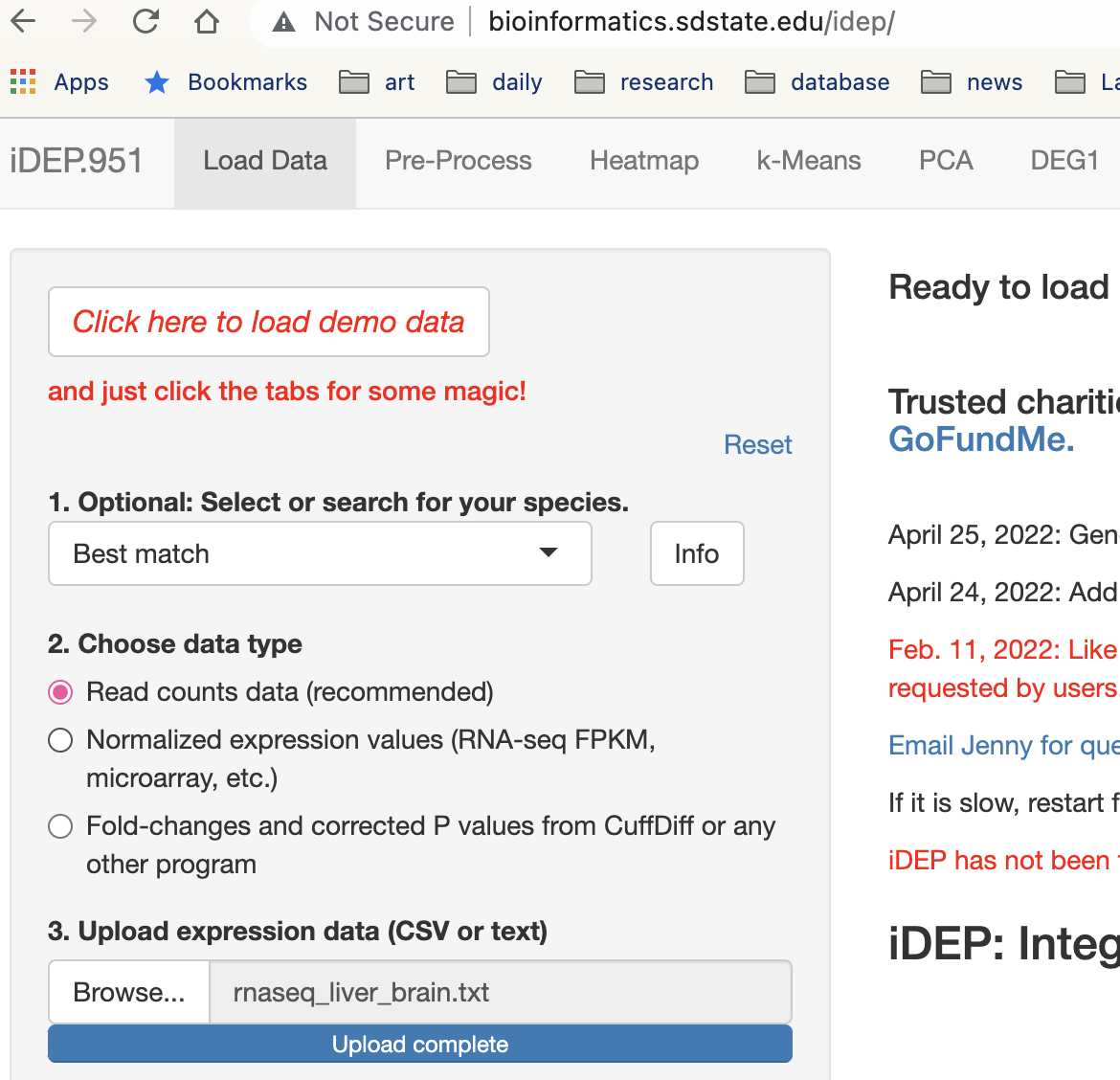
4-1. Data upload
Download the results of the entire RNAseq analysis.
https://github.com/mariesaitou/Bio326/blob/master/docs/assets/RNAseq_for_lab_2022/materials/rnaseq_liver_brain.txtAnd go to iDEP
http://bioinformatics.sdstate.edu/idep/
Upload the gene expression matrix and iDEP automatically runs (it detect the species as well)
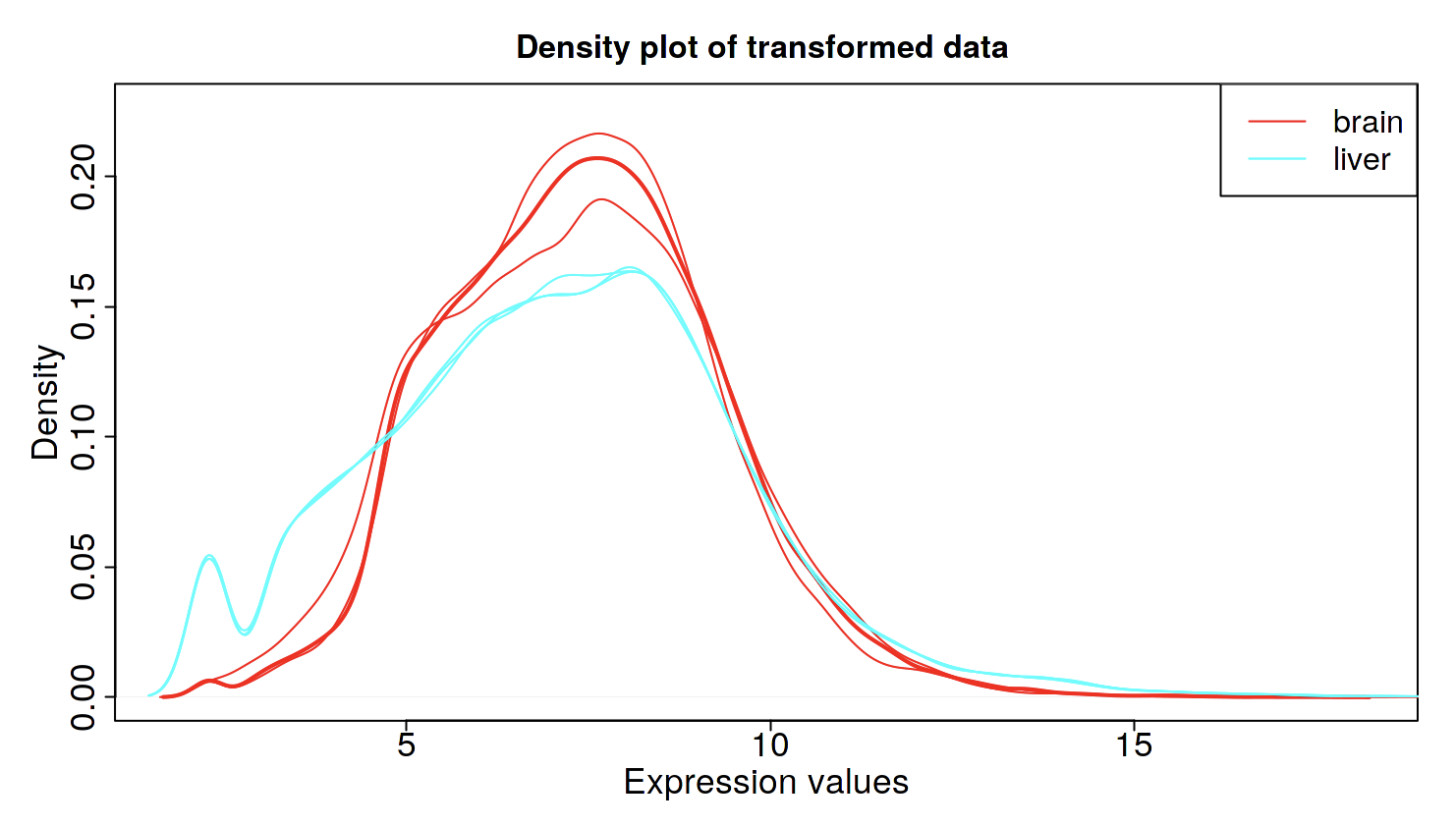
4-2. Quality check “Pre-Process”
Keep genes with minimal counts per million (CPM) in at least n libraries:
Min. CPM: 1, 2 libraries
VST: variance stabilizing transform
(I prefer to make it a bit stricter than default setting).
This is a density plot of normalized transcriptome of each sample. Brain and liver look slightly different, which makes sense. If some samples look obviously different from others (such as the expression is too high/low), it may be better to remove those samples.
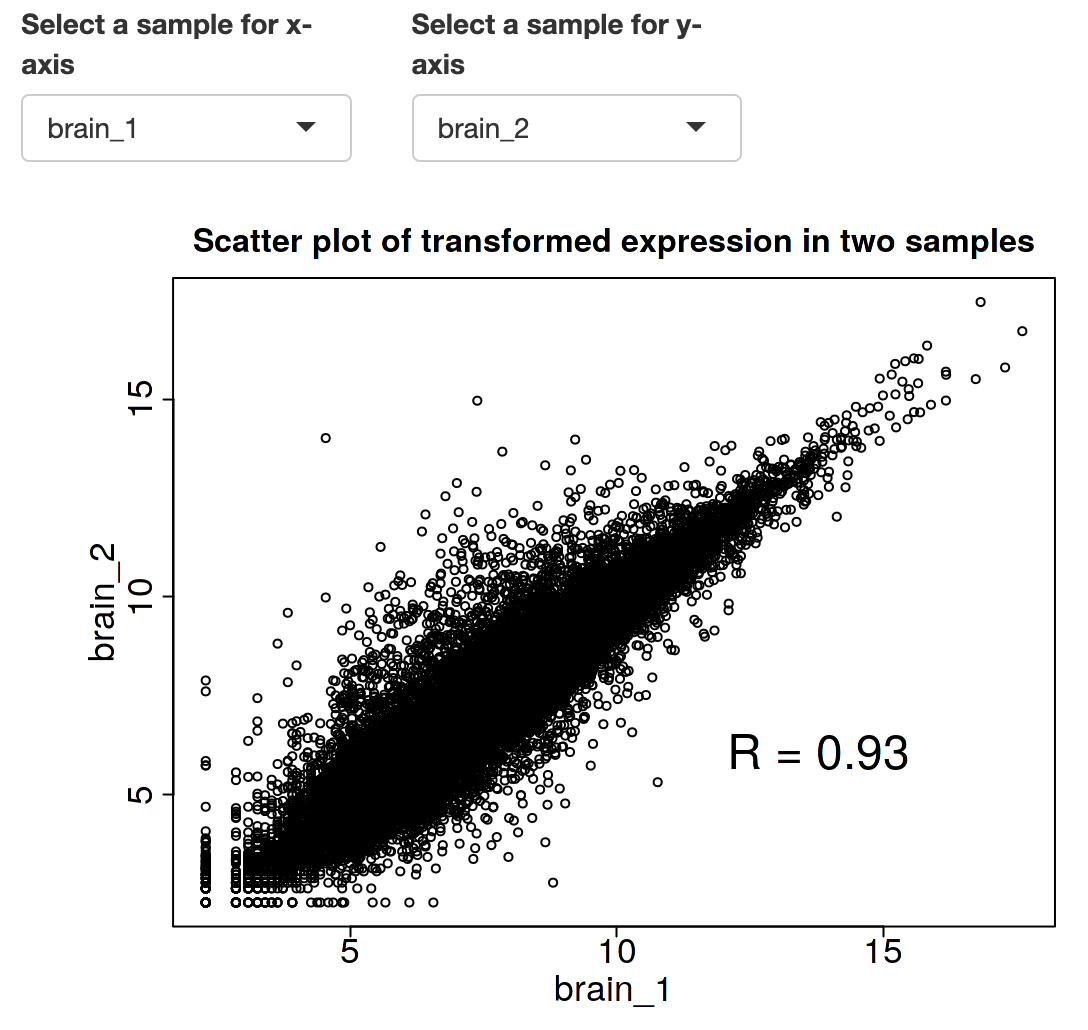
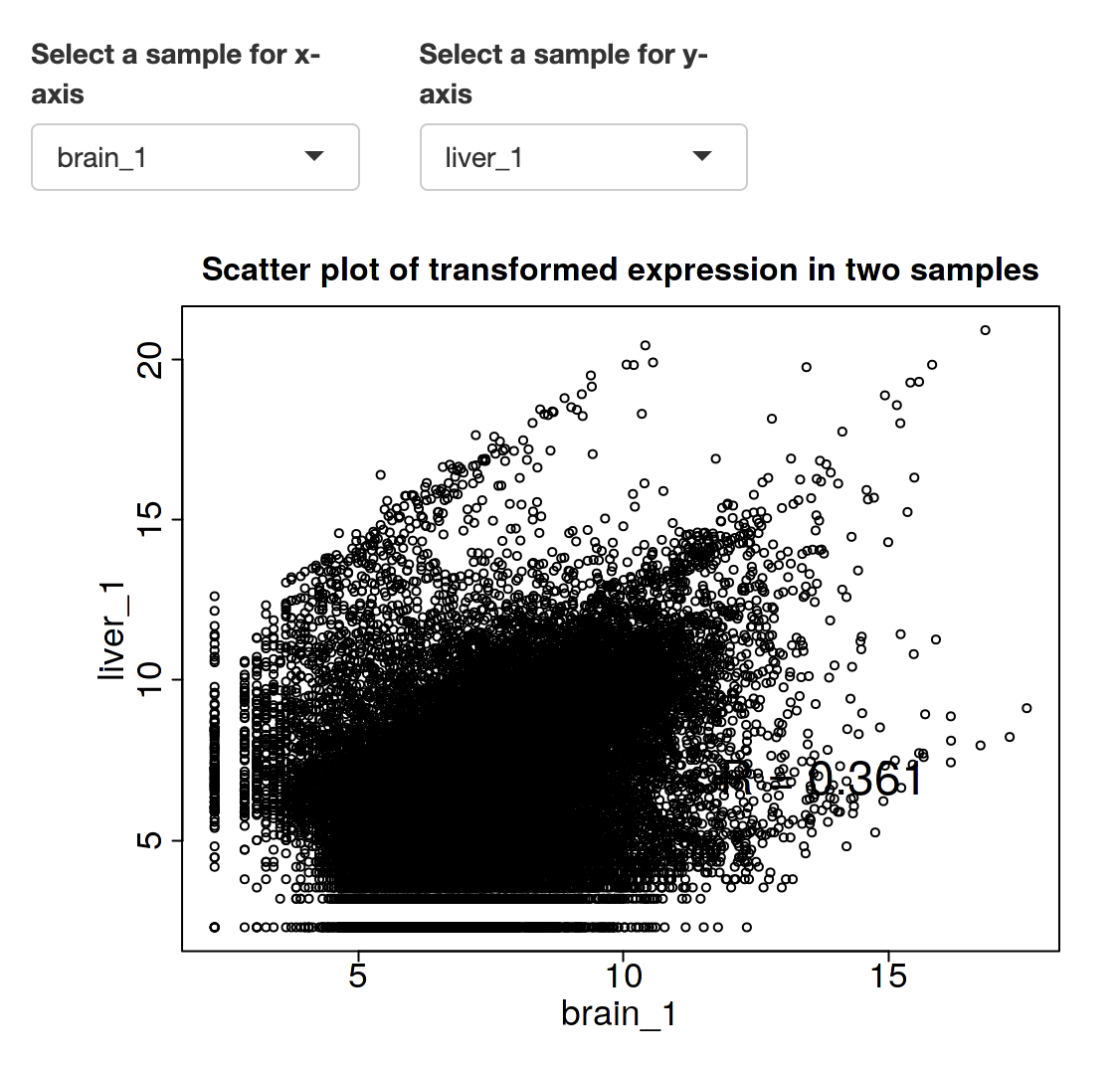
Transcriptome comparison between brain1 and brain2 samples: high correlation.
Transcriptome comparison between brain and liver samples: not very high correlation.
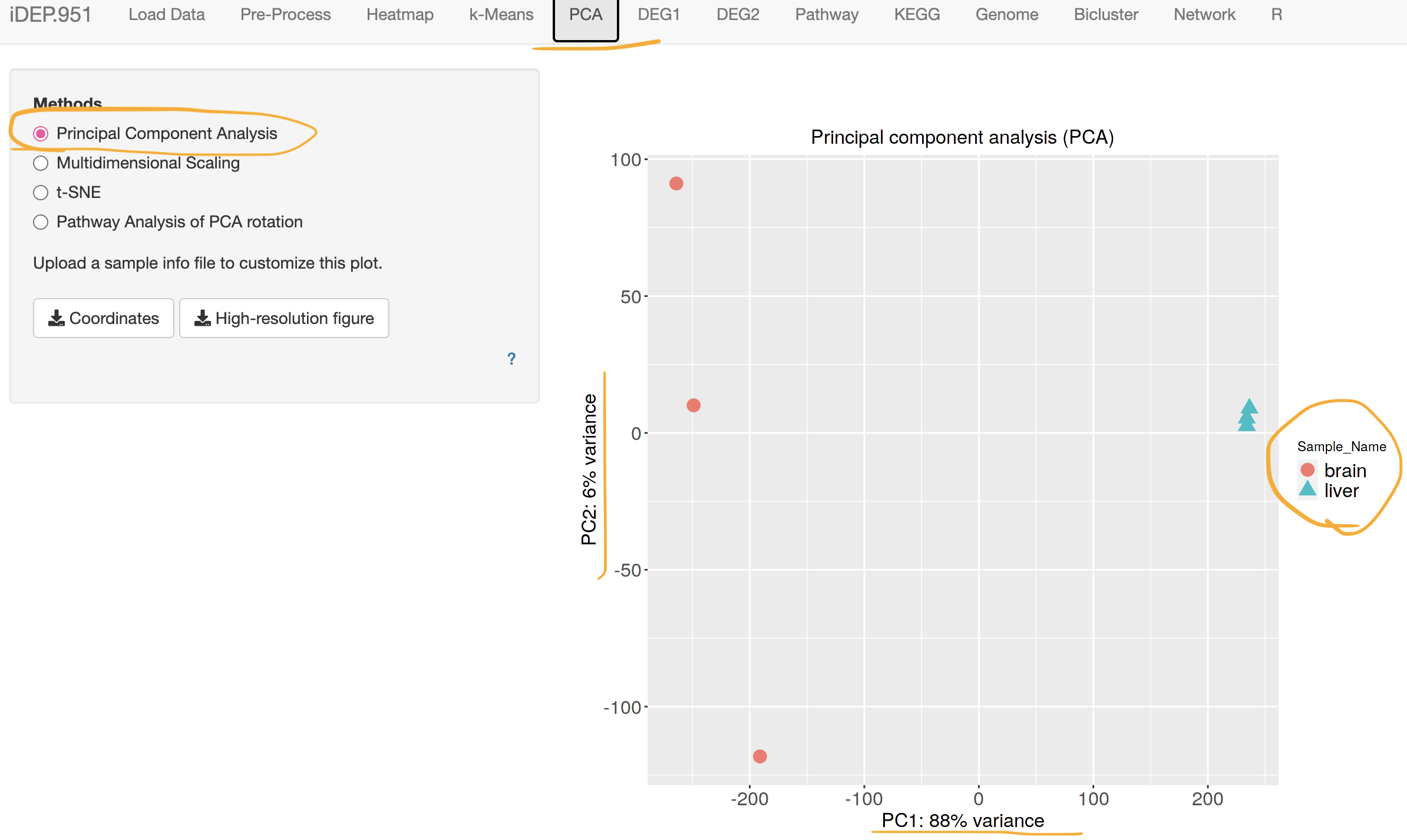
4-3.Principal component analysis (PCA)
Visualize overall trend of all the gene expression. Each dot is each sample, colored by tissue.
X axis - Principal component 1 (combination of certain gene expressions) explains 88% of the transcriptome variation across samples (mostly the difference between brain and liver).
Y axis - Principal component 2 (combination of certain gene expressions) explains 6% of the transcriptome variation across samples (mostly the difference among brain).
If one sample is obviously distant from others (=shows unusual expression patteren), it may be better to remove that sample from the analysis.
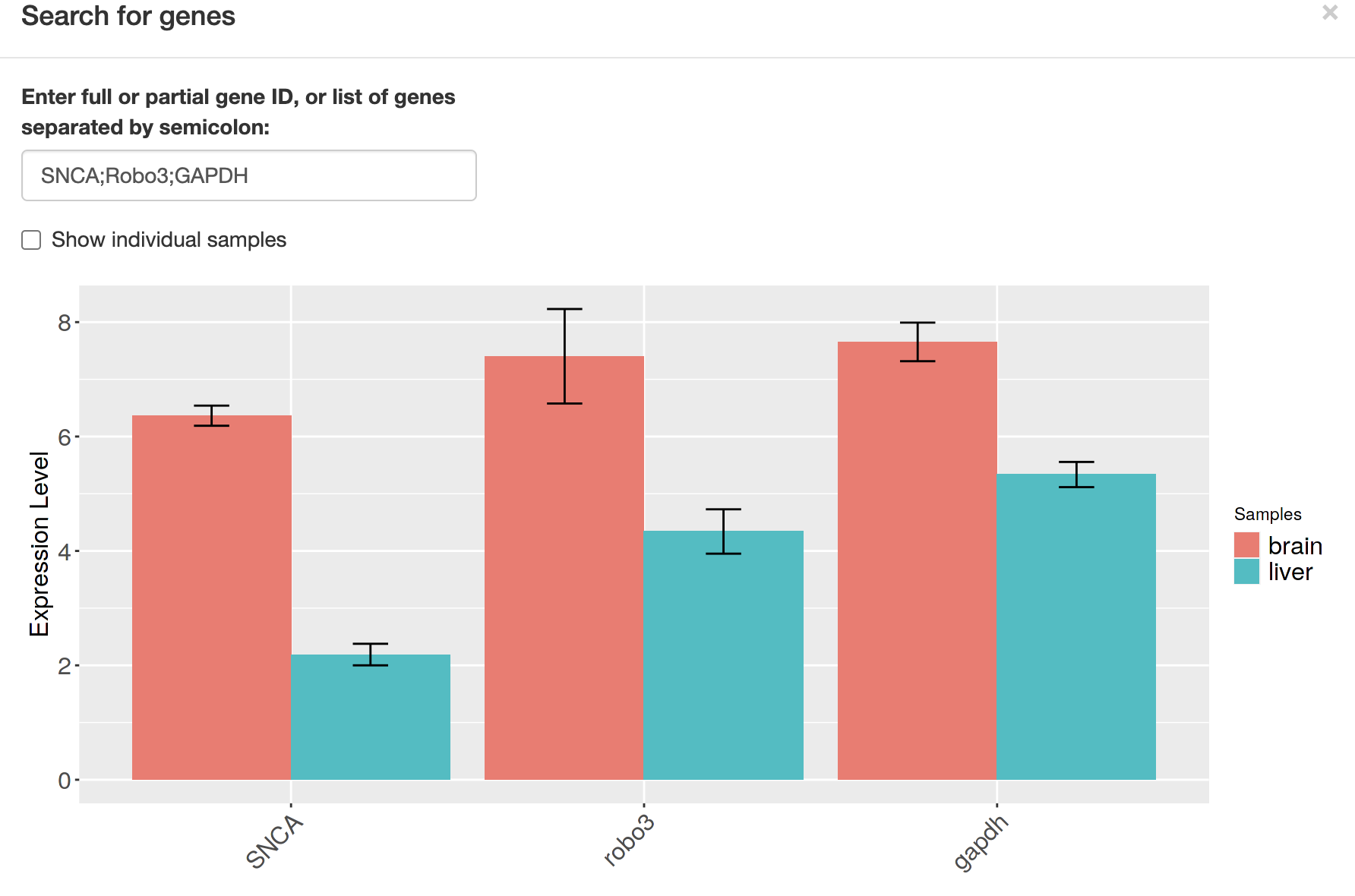
4-4. Visualize the expression of your favorite genes
“Pre-Process” -> “Plot one or more genes”
Type your favorite genes…
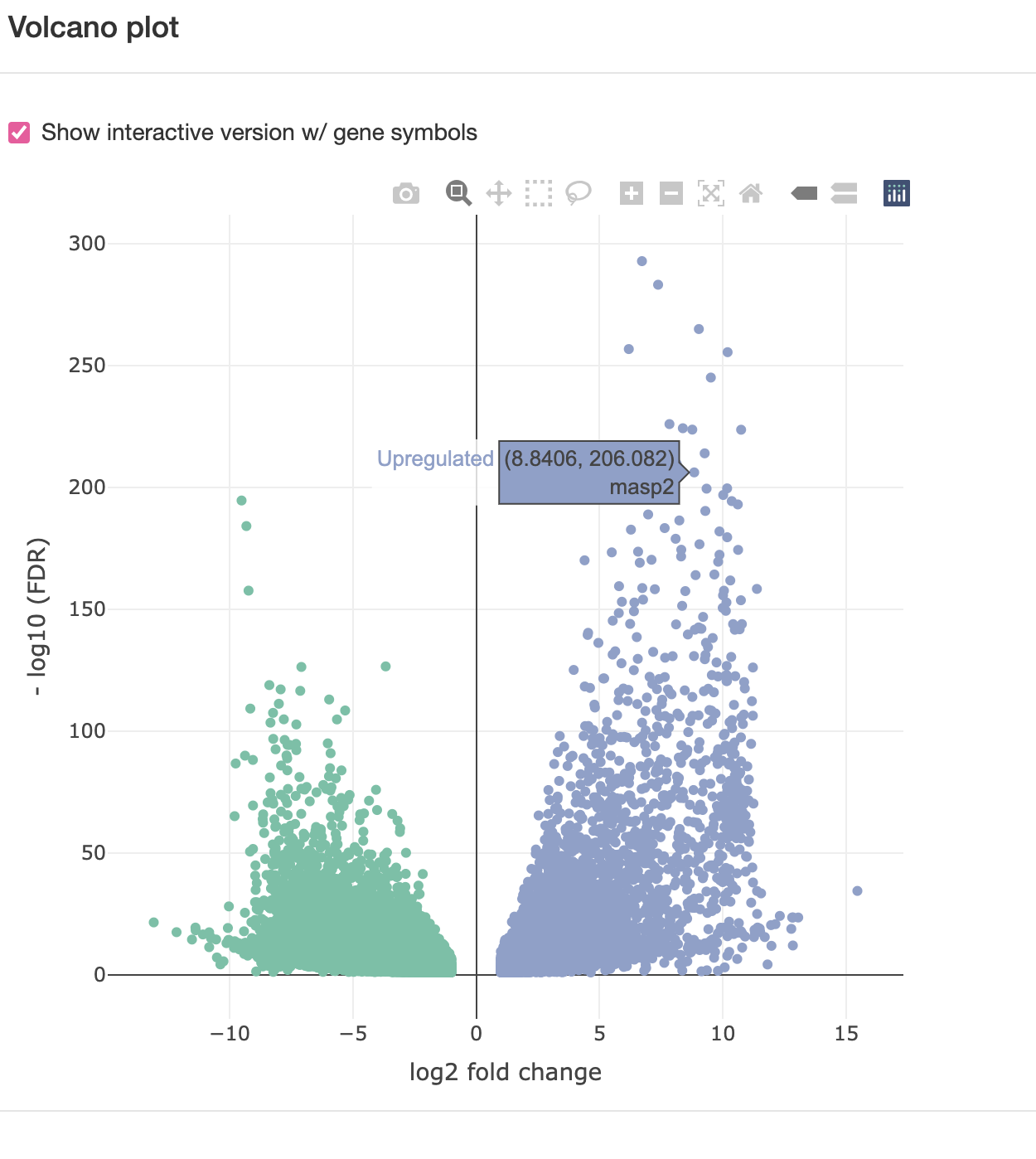
4-5. Differentially expressed genes (DEG)
Go to DEG1 tab
“DESeq2”
False discovery ratio cutoff: 0.05
Minimum fold change: 1.5
(You can download the list of deferentially expressed genes)
Go to DEG2 tab
liver-brain comparison
downregulated: more expression in brain (metabolic genes),
unregulated: more expression in liver (signaling genes)
Volcano Plot, check “Show interactive version w/ gene symbols”.
X-axis: fold change, Y-axis: false discovery ratio (statistical significance of the difference)
“Enrichment analysis for DEGs:”
Gene Ontology Analysis of differentially expressed genes 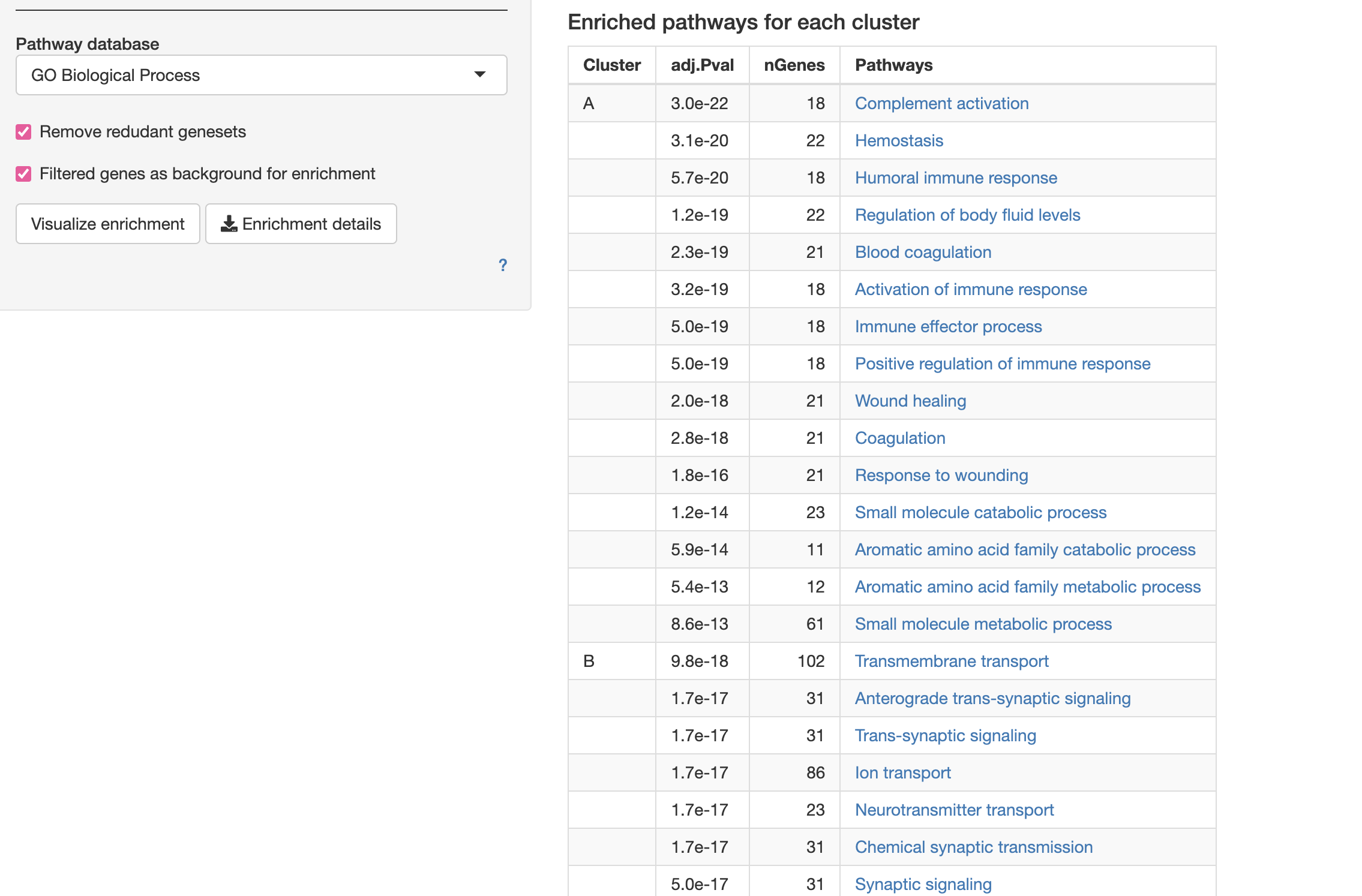 Also, do play around with Enrichment tree, Network etc.
Also, do play around with Enrichment tree, Network etc.
KEGG Analysis - if you have a particular pathway of interest.
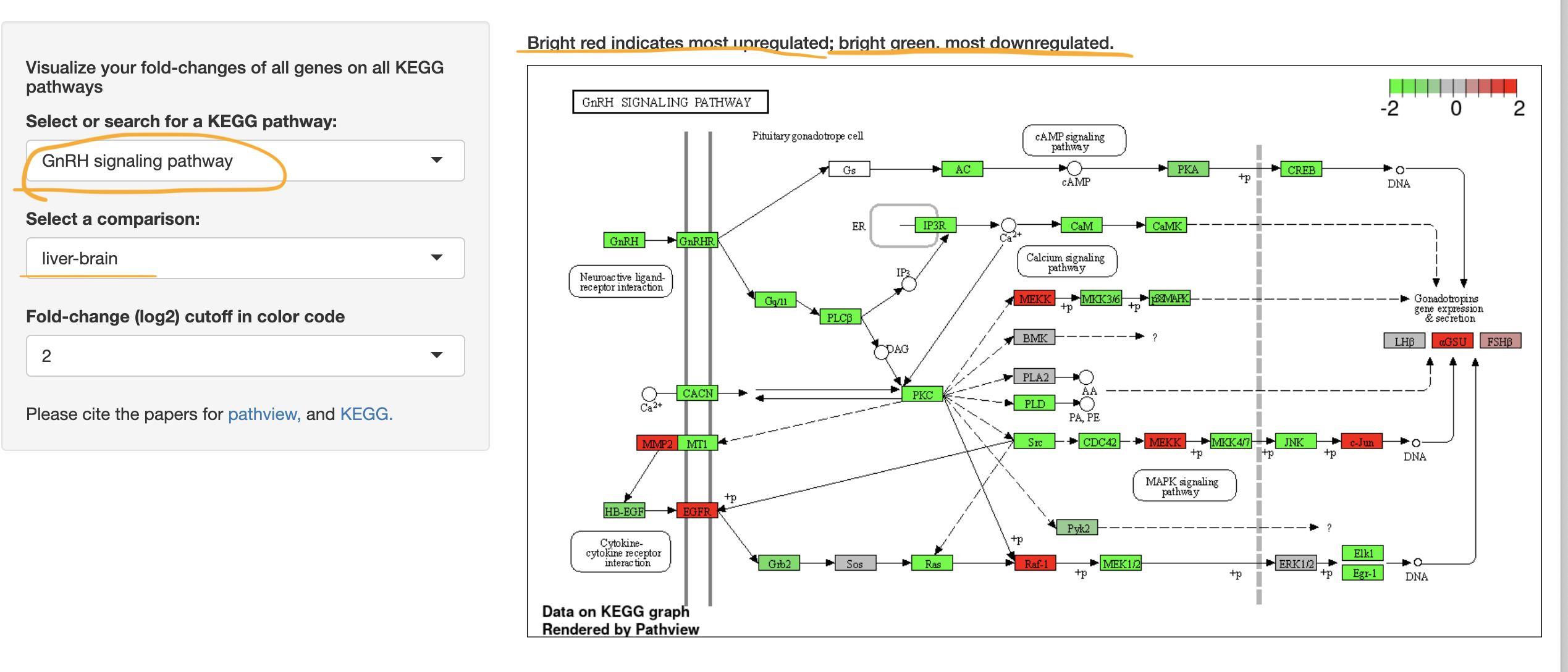
Others.
Genome: if you are interested in the distributions of the differentially expressed genes in the genome.
K-Means: if you have three or more conditions, “K-means” can classify the gene expression pattern more than up/down.
Enjoy!
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] Rcpp_1.0.8.3 rstudioapi_0.13 whisker_0.4 knitr_1.39
[5] magrittr_2.0.3 workflowr_1.7.0 R6_2.5.1 rlang_1.0.2
[9] fastmap_1.1.0 fansi_1.0.3 stringr_1.4.0 tools_4.1.2
[13] xfun_0.31 utf8_1.2.2 cli_3.3.0 git2r_0.30.1
[17] jquerylib_0.1.4 htmltools_0.5.2 ellipsis_0.3.2 rprojroot_2.0.3
[21] yaml_2.3.5 digest_0.6.29 tibble_3.1.7 lifecycle_1.0.1
[25] crayon_1.5.1 later_1.3.0 sass_0.4.1 vctrs_0.4.1
[29] promises_1.2.0.1 fs_1.5.2 glue_1.6.2 evaluate_0.15
[33] rmarkdown_2.14 stringi_1.7.6 bslib_0.3.1 compiler_4.1.2
[37] pillar_1.7.0 jsonlite_1.8.0 httpuv_1.6.5 pkgconfig_2.0.3