Bio322.ensembl1
Marie Saitou
8/23/2021
Last updated: 2021-09-13
Checks: 2 0
Knit directory: Bio322/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 42d2ed2. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Untracked files:
Untracked: Group.csv
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ensembl1.rmd) and HTML (docs/ensembl1.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 42d2ed2 | mariesaitou | 2021-09-13 | wflow_publish(“analysis/ensembl1.Rmd”) |
| html | dbbc271 | mariesaitou | 2021-09-11 | Build site. |
| Rmd | 56697ed | mariesaitou | 2021-09-11 | wflow_publish(“analysis/ensembl1.Rmd”) |
| html | bea42ca | mariesaitou | 2021-09-10 | Build site. |
| Rmd | 4cb2f58 | mariesaitou | 2021-09-10 | wflow_publish(“analysis/ensembl1.Rmd”) |
| html | b21b484 | mariesaitou | 2021-09-10 | Build site. |
| Rmd | 899b470 | mariesaitou | 2021-09-10 | wflow_publish(“analysis/ensembl1.Rmd”) |
| html | f139bdf | mariesaitou | 2021-09-10 | Build site. |
| html | f863fe0 | mariesaitou | 2021-09-10 | Build site. |
| html | c867c58 | mariesaitou | 2021-09-10 | Build site. |
| Rmd | c41e75f | mariesaitou | 2021-09-10 | wflow_publish(“analysis/ensembl1.Rmd”) |
0. Introduction
This is a set of introductory hands-on exercises to get to know more about the genomes of various animals with public databases. These exercises align the learning goals from module 1 and lectures of Bio322. https://www.nmbu.no/course/BIO322
Relevant chapters: 1, 4, and 6 of the textbook (Introduction to Genomics – 3rd edition. Oxford University Press.).
Task 4 may take some time for the server to complete the job, so you can start from 4. and can go back to other tasks while the server is processing the analysis.
You can (don’t have to) write your answers at OneNote. We may pick up some impressive answers next time!
Task 1. Human genome
Let’s explore actual genome databases and get to know the genome contents and feature.
Click the following icon. 
Here, as one example, we are going to look into BRCA2 gene.
 You can see multiple transcripts of the BRCA2 gene
You can see multiple transcripts of the BRCA2 gene
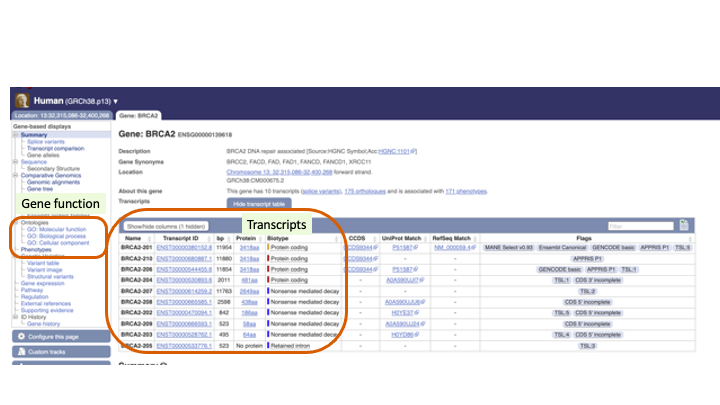
Click “Ontologies” and “Phenotypes” to explore the reported function of this gene.
Scroll down to see the transcripts and regulartory regions 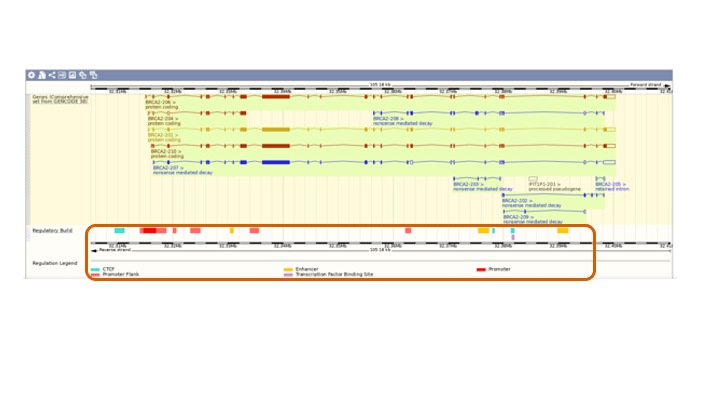
Here, there are several questions about the human genome content:
Question 1
- (1-A) What mechanism generates multiple transcripts from one gene?
- (1-B) What is an enhancer, promoter, and transcription factor binding site?
- (1-C) When you want to know function of a gene, what research plan would you make?
Task 2. Multiple genome contents
Now, we are going to compare human genome and genomes of other animals.
Go back to the human genome page, and click “(i) More information and statistics”
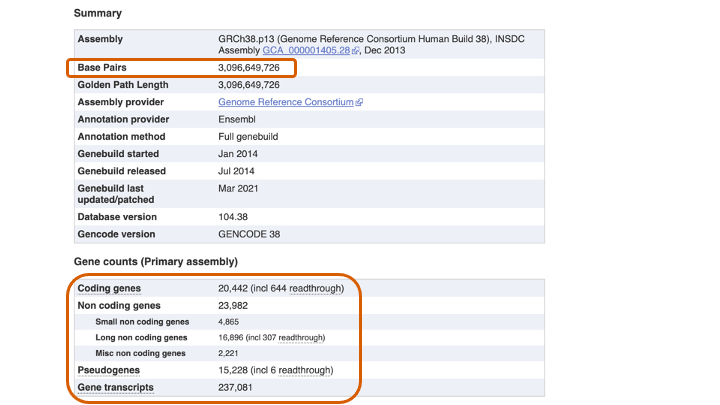
Question 2
- (2-A) How many are the Base Pairs, Coding genes, Non-coding genes, Pseudogenes, Gene transcripts in the human genome?
Similarly, let’s explore the genome of
- Chicken
- Common carp
- Saccharomyces cerevisiae (What is its common name? Please explore.)
and
(2-A, continued) write down: Base Pairs, Coding genes, Non-coding genes, Pseudogenes, Gene transcripts in their genomes.
(2-B) Why is there variation in genome size and gene numbers? How they have been changed in the process of evolution?
(2-C) How do non-coding genes work?
(2-D) What are pseudogenes? How are they generated?
(2-E) Which species has the most genes? What are the advantages of having many genes?
Task 3. Synteny
In this task, we are going to compare the structure of multiple genomes.
SynCircos is a tool to compare the contents of chromosomes from multiple species.
First, compare chromosome one of human, gorilla, mouse and cow.
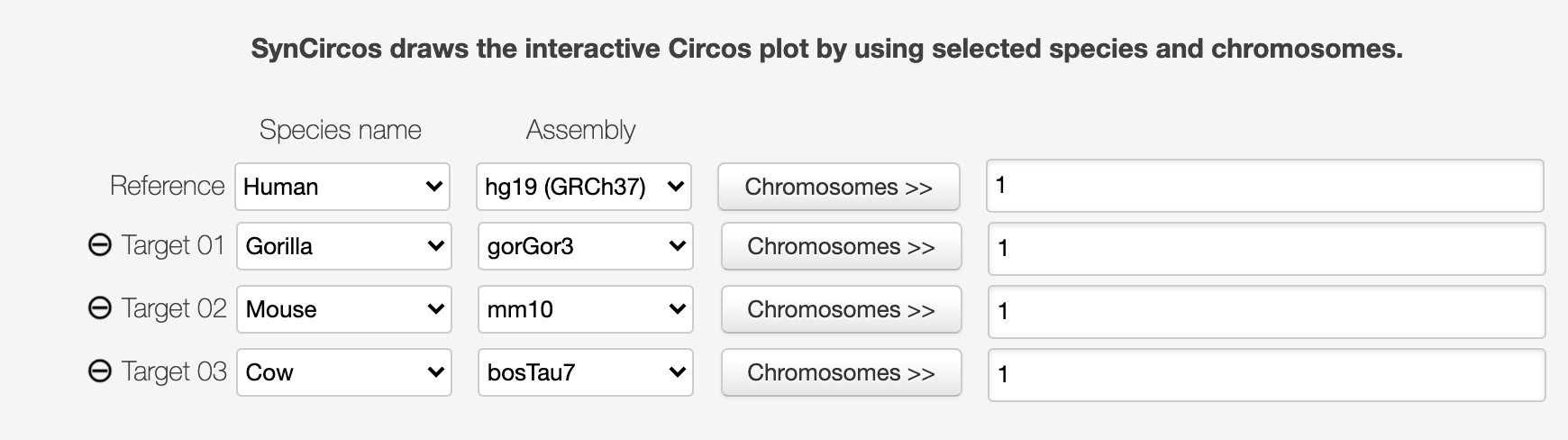
You will get a circos1 that compares chromosome one of all the four animals. We can see that human and gorilla have a quite similar chromosome one content.
However, mouse has only one-third of human chromosome one’s content, and cow’s chromosome one is entirely different from human’s.
So, where in the genome do cow and mouse have human chromosome one’s elements?
Let’s compare human chromosome one and chromosomes of mouse and cow.

And download the result. 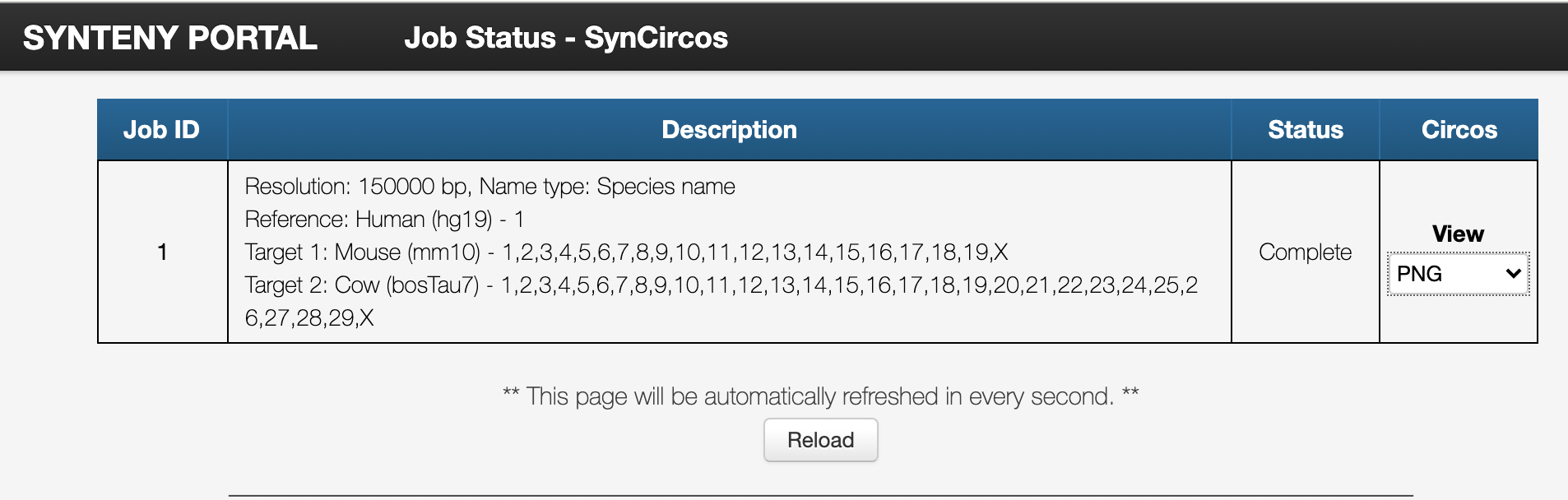
Question 3
- (3-B) Did you successfully get the result? Where in the genome do cow and mouse have human chromosome one’s elements?
Task 4. Dotplot
In this task, we are going to compare the structure of two sequences using “pipmaker”.
In our research at CIGENE, we are investigating the structure of Atlantic salmon genome. Now, we are going to compare two sequences of two individuals. You gan get the sequences here:
You can browse the content of the fasta files in TextEdit or similar software.
Let’s open PipMaker (from the above webpage) Put the first and second sequence, your email address and “Submit”. Some time after, you will receive an email with results.
Let’s open “dot.pdf”.
Question 4
What did you see?? Can you interpret it? What is happening in this genomic region?
See the instruction of pipmaker to explore the data.
Task 5. Gene gain and loss
Evolutionary analysis of tumore-suppressor genes between species. Cancer is a disease of somatic mutations, and the risk of (reviewed in Tollis et al., 2017, BMC. biol.)
Elephant has many copies of the key tumore-suppressor gene TP53 (Sulak et al., 2016, Vazquez and Lynch, 2021)
Let’s explore TP53 in various genomes. Go to https://www.ensembl.org/index.html
First, search “tp53” in the window.
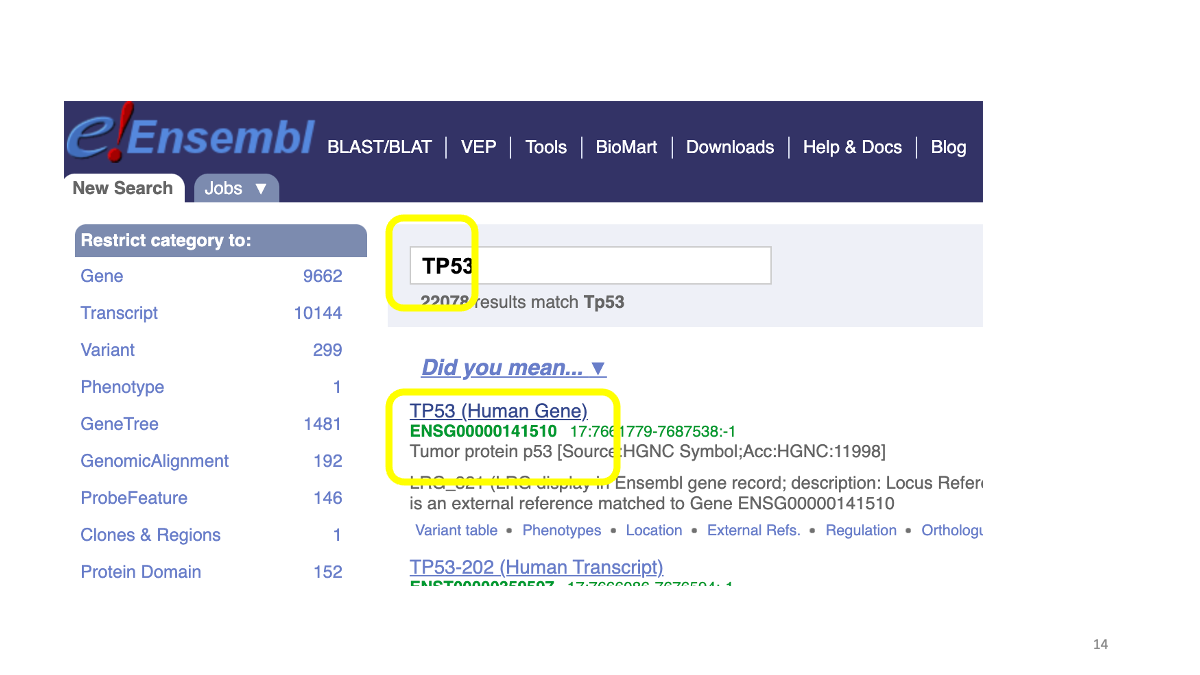
You will find human TP53 gene. On the left side, under “Comparative Genomics” tubm there is “Gene gain/loss tree” section. Let’s open this. 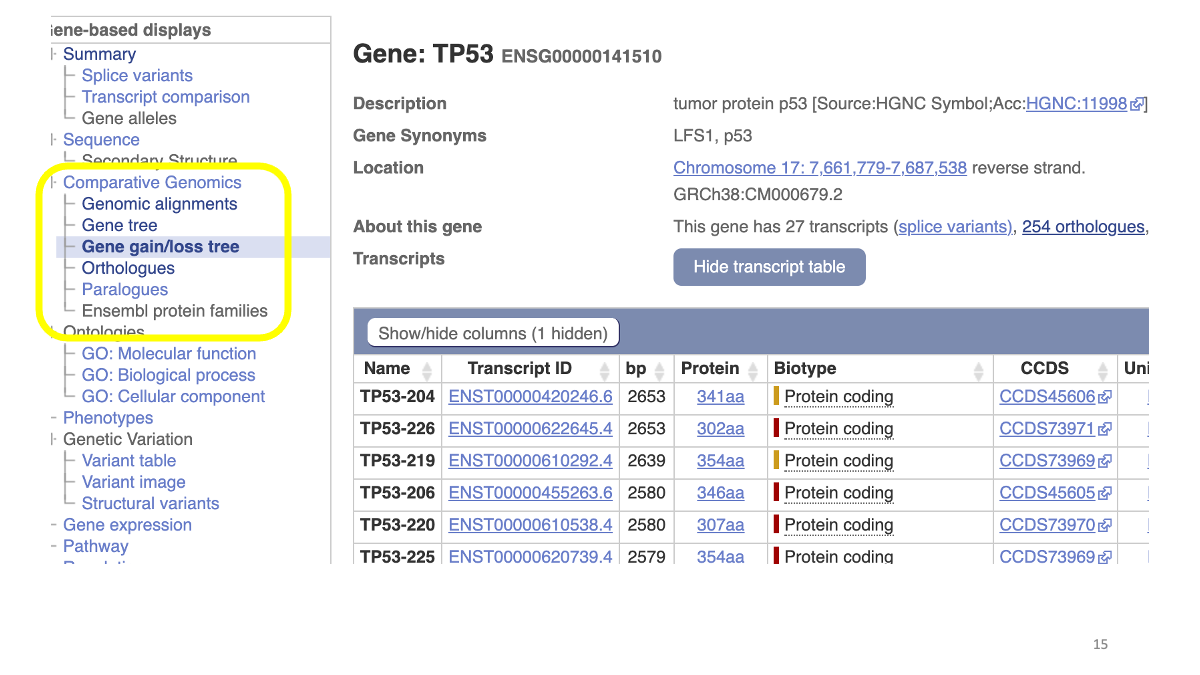
Then, you will see a phylogenetic tree of various animals, with the number of TP53 genes.You can compare gene gain/loss events in the lineages. The gene number refers to number of paralogous genes, that are originated by gene-duplication events. Humans has tree, TP53, TP63 and TP73.
You can collapse/reopen subtrees by clicking the circle and triangle.
Question 5
- (Question 5-1) Find humans, nonhuman primates, and elephant. How many TP53 genes does a elephant have?
- (Question 5-2) Find “unusual” species with many more/fewer TP53 genes compared with other species.
6. Well done!
How did you enjoy the analysis? Let us know if you have questions and comments!
Next time, we will look into individual genetic differences of human :-)