Genome Browser 2022
Marie Saitou
17/09/2022
Last updated: 2022-10-19
Checks: 2 0
Knit directory: Bio322/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version b671ebe. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: 210922_genome expression_epigenetics.pptx
Untracked: 210927.module1.2.RNAseq_on_Galaxy.pdf
Untracked: 220317_Advanced topics in genomics.docx
Untracked: 220317_Advanced topics in genomics_MarieSai.docx
Untracked: BIO322_Teaching plan BIO322 2021.docx
Untracked: Bio322.09132021.pdf
Untracked: Bio322.09132021.pptx
Untracked: Bio322.09152021.backup.pptx
Untracked: Bio322.09152021.pdf
Untracked: Bio322.09152021.pptx
Untracked: Bio322.09202021.pdf
Untracked: Bio322.09202021.pptx
Untracked: Bio322.09272021.pptx
Untracked: Bio322.09272021/
Untracked: Bio322scRNAseq.tsv
Untracked: Bio_322.docx
Untracked: Bio_322.pdf
Untracked: Galaxy1-[intestinalData.tsv].tabular
Untracked: Galaxy2.txt
Untracked: Group.csv
Untracked: analysis/Evolution_for_lab.Rmd
Untracked: analysis/_site/
Untracked: analysis/tutorial.RNAseq.foradults.xlsx
Untracked: bio322.xlsx
Untracked: bio322_2022.pptx
Untracked: chr15_inversion-v1.0.0.zip
Untracked: gene_regulation_bio322_2022.pptx
Untracked: gwassim.txt
Untracked: intestinalData.tsv
Untracked: main_workflow.ga
Untracked: markdown_test/
Untracked: mouse_intestine_scRNAseq.txt
Untracked: oharring-chr15_inversion-9615456/
Untracked: science.abg0718_data_s1_to_s8.zip
Untracked: science.abg0718_data_s1_to_s8/
Untracked: scrna_tenx.ga
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/2022_genome.function3.Rmd)
and HTML (docs/2022_genome.function3.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | b671ebe | mariesaitou | 2022-10-19 | image data set |
| html | 69adbb4 | mariesaitou | 2022-10-17 | Build site. |
| Rmd | 1ba19cd | mariesaitou | 2022-10-17 | wflow_publish("analysis/2022_genome.function3.Rmd") |
| html | 93d8ce0 | mariesaitou | 2022-10-17 | Build site. |
| Rmd | 6d520fb | mariesaitou | 2022-10-17 | wflow_publish("analysis/2022_genome.function3.Rmd") |
| html | 9896144 | mariesaitou | 2022-10-16 | Build site. |
| Rmd | a06fd2d | mariesaitou | 2022-10-16 | wflow_publish("analysis/2022_genome.function3.Rmd") |
| html | 72a15b5 | mariesaitou | 2022-10-16 | Build site. |
| Rmd | 8686188 | mariesaitou | 2022-10-16 | wflow_publish("analysis/2022_genome.function3.Rmd") |
Genome variation and function 3
Goal: Today, we learn about transcriptome, and its association with genotype.
RNAseq and eQTL - theory
RNAseq and eQTL - browse real data
Relevant review papers if you want to further learn:
1. Lecture part
2. Hands-on tasks
Now, we will investigate real transcriptome data at GTEx Portal, which concains human RNAseq date from 948 donors across 54 tissues.
Let’s explore the human ranscriptomet database GTEx Portal. In the tutorial, I did not explain everything, so please explore it yourself and get familiarised with it.
Go to “GTEx” in the top, black bar. And search for the gene “ADH1C (Alcohol Dehydrogenase 1C)” and see the results.
Questions 1
Q1-1. See “Bulk tissue gene expression”. Where is the ADH1C gene highly expressed?
Q1-2. Search for the ADH1A and ADH1B as well. Are their expression pattern different from ADH1C? How does it happen? (Remember Simen’s module1)
Q1-3. Go back to “ADH1C”. Do you see any eQTL SNPs in “Significant Single-Tissue eQTLs for ADH1C”? What is the top eQTL SNP with the lowest P-value and which tissue is it? (You can sort the variants by clicking “p-value”.)
Open the “eQTL violin plot”. Which genotype is associated with the higher expression of ADH1C?
Q1-4. Can we conclude that the eQTL (SNP-gene combination) in that tissue in Q1-3 is the most influential for ADH1C function biologically? If not, why?
Hint
The violin plot is normalized and does not reflect the absolute expression in that tissue.
Gene expression values for all samples from a given tissue were normalized using the following procedure:
Genes were selected based on expression thresholds of >0.1 TPM in at least 20% of samples and ≥6 reads in at least 20% of samples.
Expression values were normalized between samples using TMM as implemented in edgeR (Robinson & Oshlack, Genome Biology, 2010 ).
For each gene, expression values were normalized across samples using an inverse normal transform.
Q1-5. Search for “liver” in the “Significant Single-Tissue eQTLs for ADH1C”. Let’s take a look at the violin plot of the top variant, rs2173201. Which genotype is associated with the high expression of ADH1C? Can we conclude that the eQTL (SNP-gene combination) is the most influential for ADH1C function biologically? If not, why?
Q1-6. Go to PheWAS and enter the SNP ID above (rs2173201) to see if this variant is associated with any traits. What is the top traits with the lowest P-value? EA (effect allele) means allele increases the risk of the phenotype and NEA (non-effect allele) means other alleles. Which allele (nucleotide) is associated with the risk-increasing effect of the top trait?
Q1-7. Discuss how the lower expression of ADH1C (Alcohol Dehydrogenase 1C) is associated with higher alcohol intake (you don’t have to submit it).
Result 1
Click to display
https://gtexportal.org/home/gene/ADH1C
A1-1. ADH1C is highly expressed in liver, but also in colon and stomach.
A1-2. ADH1A is mainly expressed in liver, while ADH1B is highly expressed in various tissues.
A1-3. chr4_99349450_C_T_b38 (rs283415), Adipose - Subcutaneous
A1-4. TT is associated with the higher ADH1C expression in Adipose. However, The ADH1C expression in Adipose is much lower (Median TPM = 14.23) compared to liver (Median TPM = 273.7), and we cannot conclude that the eQTL in Adipose has the primary effect for ADH1C function.
A1-5. AA genotype is associated with the higher ADH1C expression in liver. However, eQTL only indicates the statistical association between genotypes and gene expression, and this does not tell the biological mechanism regarding it.
A1-6. C allele is associated with higher alcohol intake and higher Alcohol dependency risk.
A1-7. Let’s review the results so far.
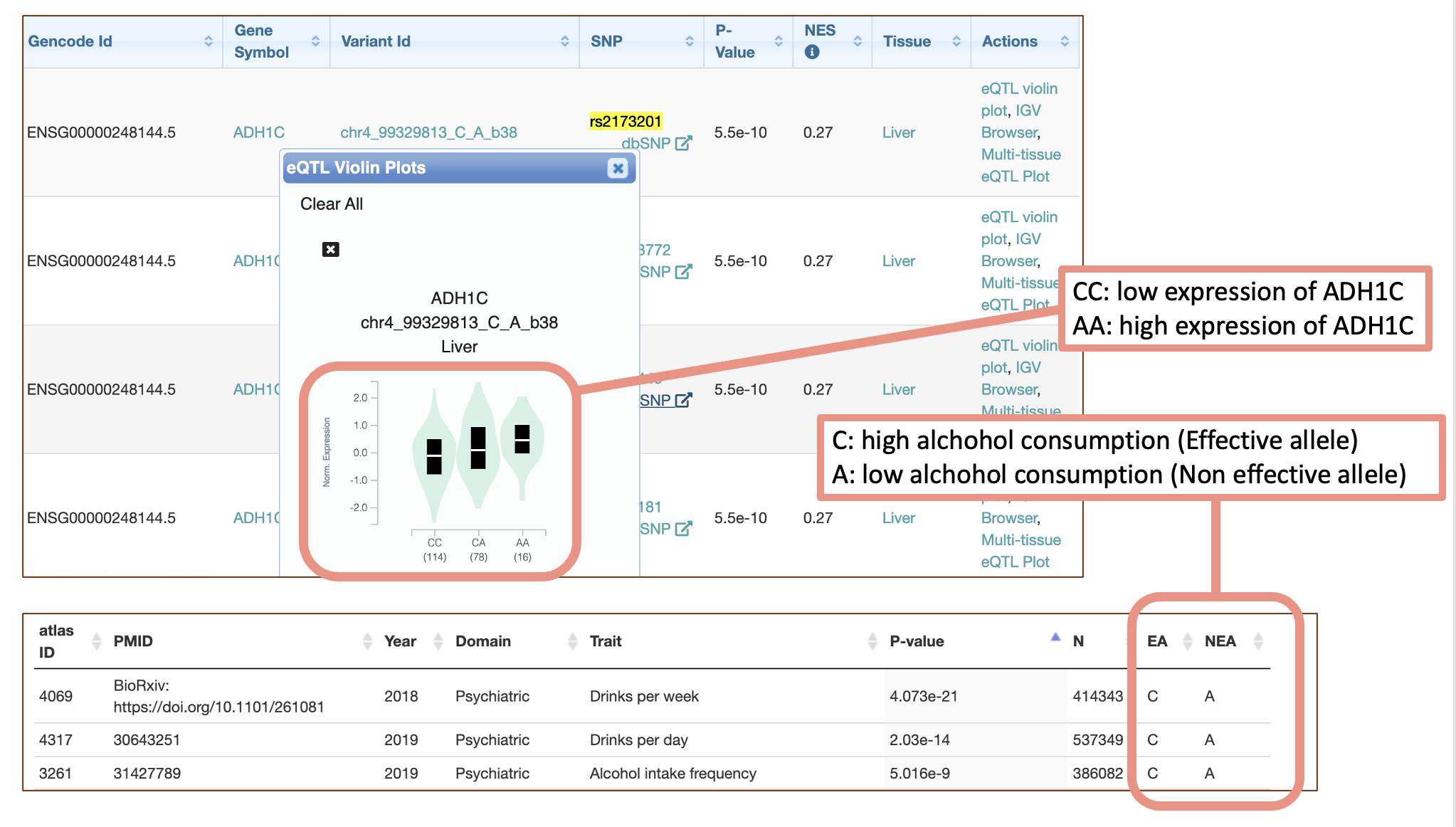
The C allele is associated with low expression of ADH1C and high alcohol consumption. One hypothesis: From the reading material, the high ADH activity tend to generate higher amount of Acetaldehyde from Ethanol, which may make that person feel sick/dissy. This makes that person uncomfortable, and make them refrain from heavy alcohol consumption.