GWAS
Marie Saitou
Last updated: 2024-10-08
Checks: 2 0
Knit directory: Bio322/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 4d446e2. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: 210922_genome expression_epigenetics.pptx
Untracked: 210927.module1.2.RNAseq_on_Galaxy.pdf
Untracked: 220317_Advanced topics in genomics.docx
Untracked: 220317_Advanced topics in genomics_MarieSai.docx
Untracked: BIO322.question.docx
Untracked: BIO322_Teaching plan BIO322 2021.docx
Untracked: Bio322.09132021.pdf
Untracked: Bio322.09132021.pptx
Untracked: Bio322.09152021.backup.pptx
Untracked: Bio322.09152021.pdf
Untracked: Bio322.09152021.pptx
Untracked: Bio322.09202021.pdf
Untracked: Bio322.09202021.pptx
Untracked: Bio322.09272021.pptx
Untracked: Bio322.09272021/
Untracked: Bio322scRNAseq.tsv
Untracked: Bio_322.docx
Untracked: Bio_322.pdf
Untracked: Galaxy1-[intestinalData.tsv].tabular
Untracked: Galaxy2.txt
Untracked: Group.csv
Untracked: analysis/Evolution_for_lab.Rmd
Untracked: analysis/_site/
Untracked: analysis/tutorial.RNAseq.foradults.xlsx
Untracked: bio322.xlsx
Untracked: bio322_2022.pptx
Untracked: chr15_inversion-v1.0.0.zip
Untracked: gene_regulation_bio322_2022.pptx
Untracked: gwassim.txt
Untracked: intestinalData.tsv
Untracked: main_workflow.ga
Untracked: markdown_test/
Untracked: mouse_intestine_scRNAseq.txt
Untracked: oharring-chr15_inversion-9615456/
Untracked: science.abg0718_data_s1_to_s8.zip
Untracked: science.abg0718_data_s1_to_s8/
Untracked: scrna_tenx.ga
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/2022_genome.function2.Rmd)
and HTML (docs/2022_genome.function2.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 4d446e2 | mariesaitou | 2024-10-08 | wflow_publish("analysis/2022_genome.function2.Rmd") |
| html | bf4d4d2 | mariesaitou | 2022-10-10 | Build site. |
| Rmd | 3d7a0e9 | mariesaitou | 2022-10-10 | wflow_publish("analysis/2022_genome.function2.Rmd") |
| html | 82d7bc2 | mariesaitou | 2022-10-10 | Build site. |
| Rmd | d99cec7 | mariesaitou | 2022-10-10 | wflow_publish("analysis/2022_genome.function2.Rmd") |
| html | 1378cc5 | mariesaitou | 2022-10-09 | Build site. |
| html | 03bf4d7 | mariesaitou | 2022-10-09 | Build site. |
| Rmd | 331672b | mariesaitou | 2022-10-09 | wflow_publish("analysis/2022_genome.function2.Rmd") |
| html | e43ff32 | mariesaitou | 2022-10-09 | Build site. |
| Rmd | 861f5c8 | mariesaitou | 2022-10-09 | wflow_publish("analysis/2022_genome.function2.Rmd") |
| html | a0cae87 | mariesaitou | 2022-10-09 | Build site. |
| Rmd | 14a66d0 | mariesaitou | 2022-10-09 | wflow_publish("analysis/2022_genome.function2.Rmd") |
| html | e507e70 | mariesaitou | 2022-10-09 | Build site. |
| html | 0c87f62 | mariesaitou | 2022-10-09 | Build site. |
| Rmd | 59378af | mariesaitou | 2022-10-09 | wflow_publish("analysis/2022_genome.function2.Rmd") |
| html | 664b5c6 | mariesaitou | 2022-10-09 | Build site. |
| Rmd | b56d9ff | mariesaitou | 2022-10-09 | wflow_publish("analysis/2022_genome.function2.Rmd") |
Genome variation and function
Goal: Today, we learn about how to analyze genetic variants and its association with phenotype.
Genome wide association study - theory
Genome wide association study - browse real data
Relevant review papers if you want to further learn…:
Depends on your interest:
Hands-on tasks
Now, we will investigate real GWAS data at GWAS ATLAS, which concains 4,756 human GWAS across 3,302 traits.
Let’s explore the human GWAS database GWAS ATLAS. In the tutorial, I did not explain everything, so please explore it and get familiarised yourselfwith it.
Case study 1
Go to “Browse GWAS” in the top, black bar. And search for the trait about hair morphology and see the results.
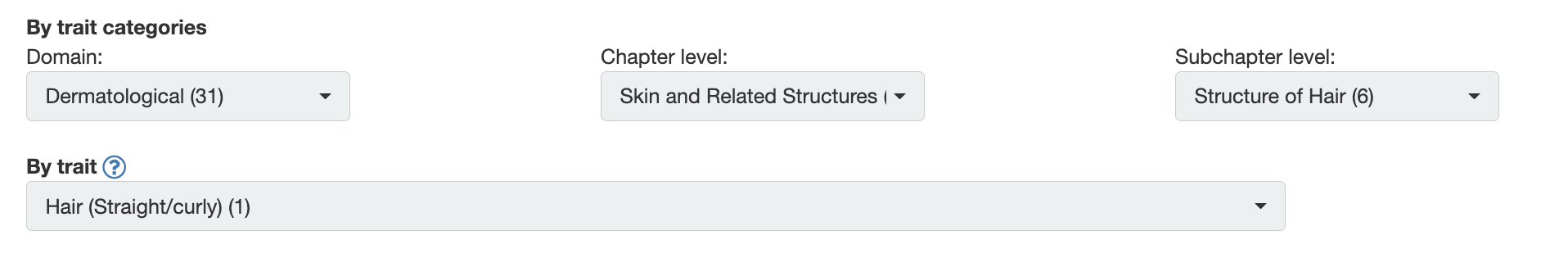
Questions 1
Q1-1. Examine the Manhattan plot. Where do you see the peak? How can we interpret this?
Q1-2. See the left table. How many individuals were investigated?
Q1-3. How many variants were investigated?
Q1-4. See the table under the Manhattan plot with gene annotations. What is the ID of top-associated SNP? (SNP ID: rsXXXXX)
Q2-5. Go to GGV map browser and enter the SNP ID above to see the global distribution of this variant. “A” is the straight hair allele, and “G” is the curly hair allele. What can we estimate from this map? Why did it happen?
Result 1
Click to display
https://atlas.ctglab.nl/traitDB/4023
A1-1. There is one big peak at chromosome 2. It is plausible that one genetic factor with very strong effect is associated with this trait.
A1-2. 4878 individuals
A1-3. 560921 SNPs
A1-4. rs260643
A1-5. Straight hair type is thought to be adaptive in East Asia in human evolution. Why? There are a lot of theory but there is not a concrete consensus yet.
if you are interested In this paper, they generated a knock-in mouse model to investigate the function of hair-morphology altering variant. In the mouce model, they observed increased hair thickness, but also change the morphology of mammary and eccrine glands.
Case study 2
Go to “Browse GWAS” in the top, black bar. And search for the trait about eyesight and see the results.

Questions 2
Q2-1. Examine the Manhattan plot. Where do you see the peak? How can we interpret this?
Q2-2. See the left table. How many individuals were investigated?
Q2-3. How many variants were investigated?
Q2-4. See the bottom Manhattan plot with gene annotations. What is the top-associated gene?
Q2-5. (Discussion topic) - Do some literature search about the top-associated genes and assume how this gene is associated with the trait, “Reason for glasses/contact lenses: For short-sightedness”. To further understand the mechanism, what experiment would you plan?
Result 2
Click to display
https://atlas.ctglab.nl/traitDB/3539
A2-1. There are multiple peaks, in the chromosomes 1, 2, 4, 6, 8, 10, 15… The obserbation implies that multiple loci are associated with this trait (The trait is polygenic).
A2-2. 78647 samples (See the left box, “N”)
A2-3. 9223534 SNPs (See the left box, “Nsnps”)
A2-4. PRSS56 (the dot with lowest P-value = highest -log10P value)
A2-5. Put what you have discussed on CANVAS “Discussion” with the full names of participants who contributed to the discussion.
Case study 3 (You have to submit only Case study 3)
Go to “Browse GWAS” in the top, black bar. And search for a trait you like,and examine and inter the results.